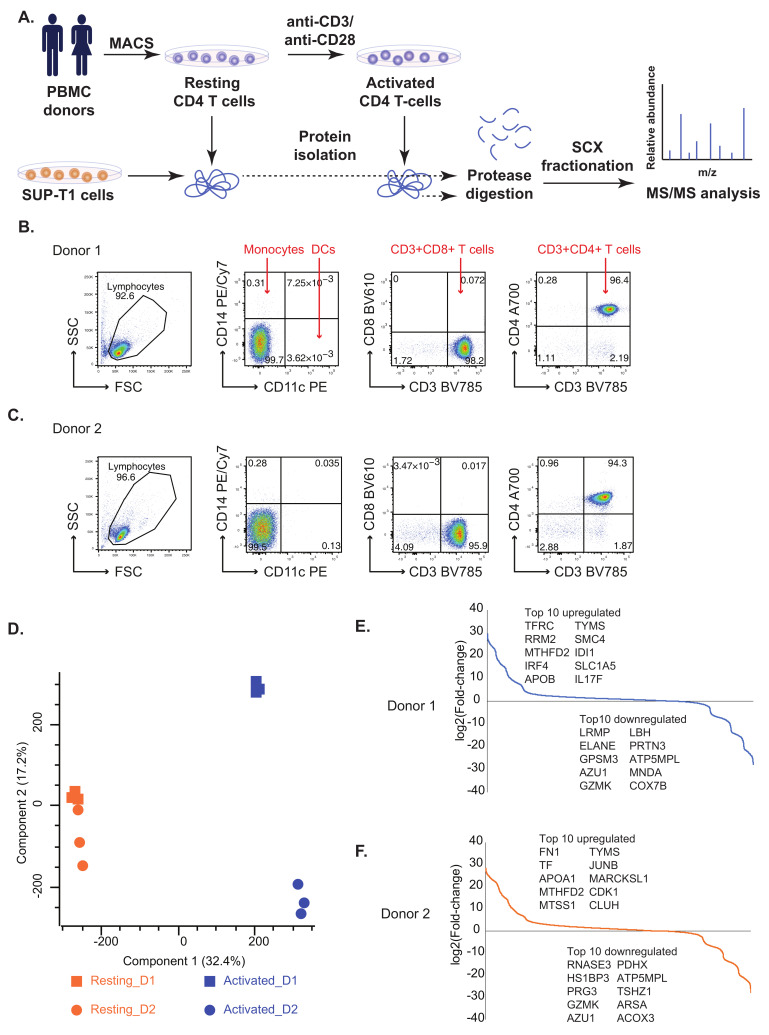
Figure 1.
(A) A workflow for the comparative proteomics analysis of resting and activated primary human CD4+ T cells as well as SUP-T1 lymphoma T cells. Primary human CD4+ T cells were purified using magnetic beads (MACS cell separation) from PMBCs of healthy donors. Resting (unstimulated) CD4+ T cell samples were harvested (washed and shock-frozen) for protein extraction immediately after isolation. A fraction (1 × 107) CD4+ T cells were activated for 72 h by anti-CD3/anti-CD28 stimulation before cells were harvested/shock-frozen for protein isolation. SUP-T1 cells were harvested from a cell line tissue culture. Resting CD4+ T cells, activated CD4+ T cells and SUP-T1 cell samples were subjected to protein extraction. Proteins were subjected to tryptic digestion. Peptide samples obtained were subjected to strong cation exchange chromatography followed by MS/MS analysis. (B,C) Flow cytometric purity analysis of CD4+ T cell preparations for Donor 1 (B) and Donor 2 (C). Purified CD4+ T cells were stained with fluorescent antibodies for CD11c, CD14, CD3, CD8 and CD4. Both preparations contained >94% CD3+CD4+ T cells and (in total) less than 0.5% contaminating CD11c+, CD14+ or CD8+ cells. (D) Principal Component Analysis (PCA) plot depicting common proteomic patterns in resting/resting CD4+ T cells and activated CD4+ T cells. (E,F) S-curve graphs showing the distribution of fold-changes in Donors 1 and 2 and the top differentially expressed proteins.