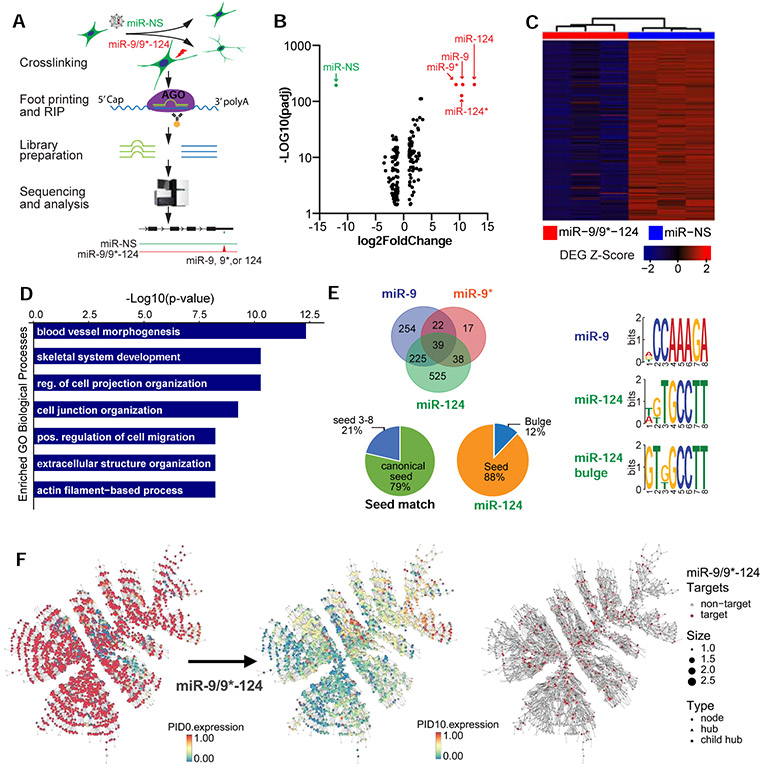
Figure 4. Targets of miR-9/9*-124 are enriched in non-neuronal gene networks.
(A) Overview of AGO-HITS-CLIP to identify targets of miR-9/9*-124 during neuronal conversion. (B) Aggregate analysis of miR-9/9*-124 and miR-NS conditions shows that miR-9, miR-9*, miR-124, and miR-124* (red) were enriched in RISC complex immunoprecipitated with AGO antibody relative to total input in contrast to the control miR-NS (green). (C) Heatmap of genes downregulated in miR-9/9*-124 condition over miR-NS at PID 7. Signal intensity is based on normalized CPM values, and data is shown as a z-score normalized log2CPM. (D) Top GO terms corresponding to downregulated miR-9/9*-124-target genes at PID 7. (E) RNAhybrid analysis of miR-9/9*-124 targets detected by AGO-HITS-CLIP (left). Various modes of seed positions on miRNAs were detected to pair with enriched peaks. Canonical seed = seed 1-6 or 2-7 of miRNAs. The miRNA binding motif analysis for miR-9, miR-124 seed-matches, and miR-124 bulge detected by MEME motif analysis. (F) Comparative gene expression analysis (scaled 0-1) of the top fibroblast-enriched network between days 0 and 10 of reprogramming. The fibroblast network is significantly enriched for targets of miR-9, miR-9*, and miR-124 as identified by AGO-HITS-CLIP (red).