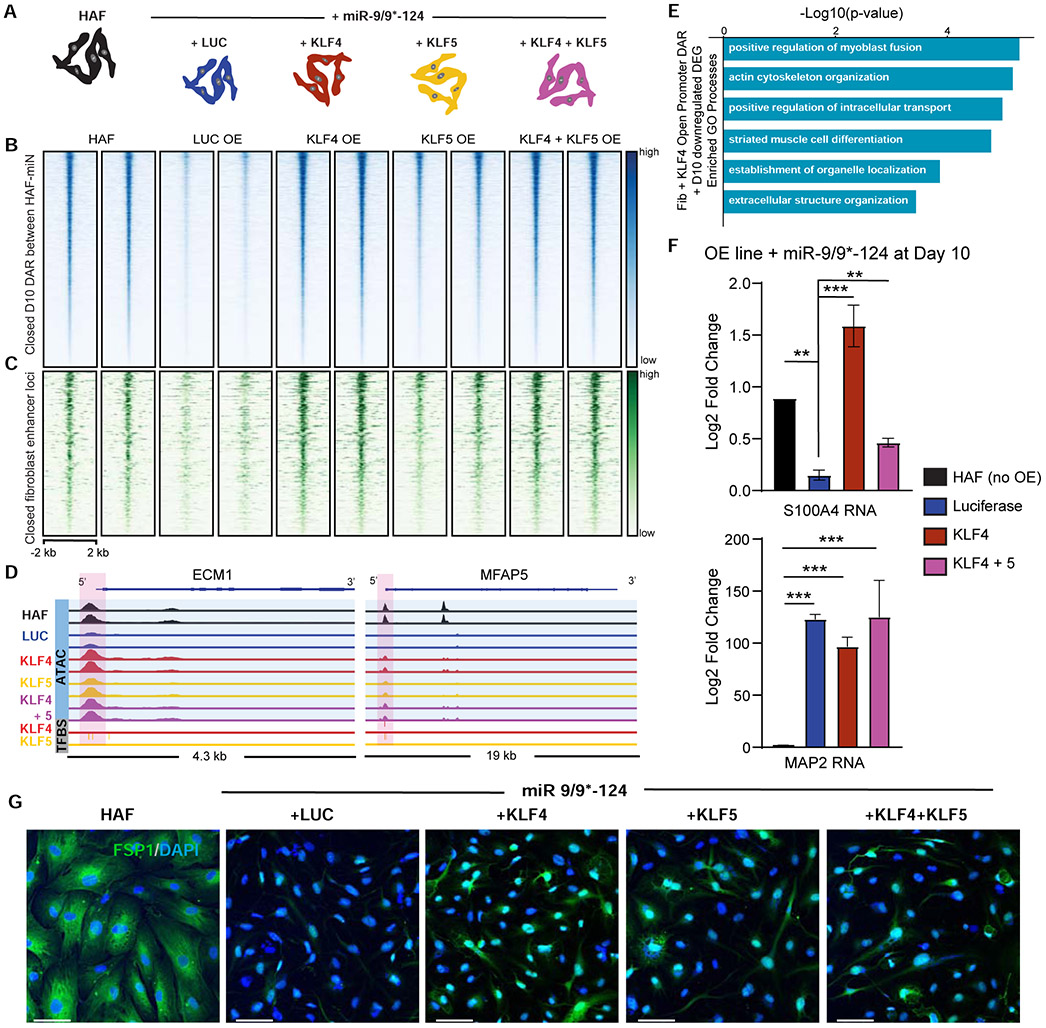
Figure 6. miR-9/9*-124 repression of KLF4 and KLF5 is required for complete erasure of fibroblast identity.
(A) Experimental conditions for studying the role of KLF4 and KLF5 in miR-9/9*-124 reprogramming. (B) Heatmaps of ATAC-seq signal intensity mapped to DAR closed between HAF and miR-9/9*-124 conditions by PID 10 of reprogramming (n = 9,854). (C) Heatmaps of ATAC-seq signal intensity mapped to DAR closed between HAF and miR-9/9*-124 conditions by day 10 of reprogramming that overlap with fibroblast enhancer loci (n = 469). D) IGV snapshot showing ATAC-seq read coverage (blue) and KLF4/KLF5 transcription factor binding sites (grey) for the fibroblast-associated genes ECM1 and MFAP5. The LUC compared to KLF4 DAR peaks are highlighted in pink. (E) Top GO biological processes associated with downregulated DEGs in response to miR-9/9*-124, containing DARs that fail to close with prolonged expression of KLF4 compared to LUC control expression in day 10 of miRNA expression. (F) qPCR results for fibroblast (S100A4) and neuronal (MAP2) gene expression for LUC or KLF overexpression with miR-9/9*-124 and HAF controls at PID 10 (**p <0.01, *** p < 0.001). Error bars are s.e.m. (G) Representative images of HAFs expressing miR-9/9*-124 with LUC, KLF4, KLF5, or KLF4 and KLF5 at Day 10 immunostained for FSP1 (red) and DAPI (blue). Scale bars, 60 μm.