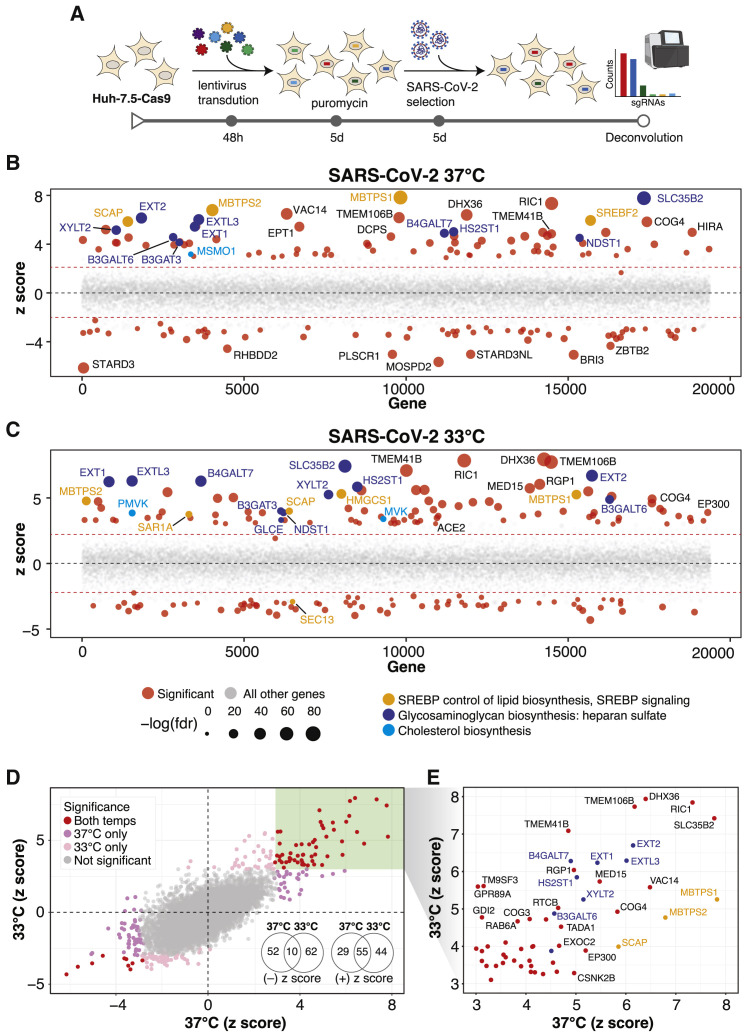
Figure 1.
Genome-wide CRISPR Screens Identify Host Factors Required for SARS-CoV-2 Infection
(A) Genome-wide CRISPR screening workflow. Cas9-expressing Huh-7.5 cells are transduced with the Brunello genome-wide CRISPR library, selected with puromycin, and infected with SARS-CoV-2 or one of three seasonal CoVs (HCoV-OC43, HCoV-NL63, or HCoV-229E). Surviving cells and mock controls are then harvested, and sgRNA abundance is determined using next-generation sequencing.
(B) Bubble plot of data from SARS-CoV-2 screens at 37°C. Red lines denote z = ±2.
(C) Bubble plot of data from SARS-CoV-2 screens at 33°C. Red lines denote z = ±2.
(D) Scatterplot comparing Z scores from (B) and (C) for SARS-CoV-2 screens at 37°C and 33°C, respectively.
(E) Subset of significantly enriched genes from SARS-CoV-2 screens at 37°C and 33°C.