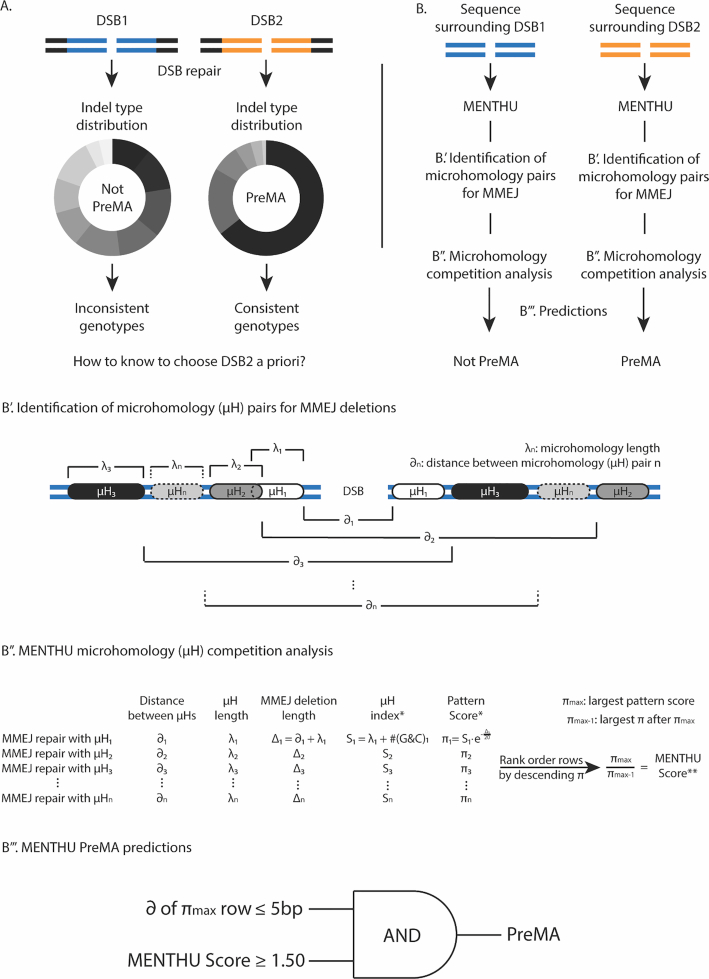
Figure 1.
MENTHU predicts which DNA double-strand breaks likely result in single majority deletions. (A) Different DNA double-strand breaks (DSBs) can generate indel profiles with dissimilar distributions. Being able to discern the genotype heterogeneity level between targetable DSBs prior to experimental applications would be beneficial for reverse genetics and gene therapy applications. (B) MENTHU (22) is a software tool that analyzes the DNA sequence surrounding any given DSB and predicts whether it will result in a PreMA: an MMEJ-mediated repaired sequence where half or more of the repair outcomes share the same genotype. B’. MENTHU identifies every possible μH pair (with homology arms μH1 to μHn of length λ1 to λn) and calculates the corresponding distance between the μHs of each pair (∂1 to ∂n). B’’. Based on the expected MMEJ deletion pattern, ∂i and λi are used to calculate the expected deletion length Δi. Pattern scores πi for every possible MMEJ deletion are calculated as described by Bae et al. (16). The MMEJ deletions are then rank ordered by descending pattern score and a MENTHU Score for the DSB is calculated by taking the ratio between the largest πmax and the second largest pattern score πmax-1. B’’’. MENTHU utilizes two criteria that need to be concomitantly true for a DSB to be labeled as a PreMA. The ∂ of the MMEJ-deletion with the highest pattern score πmax and the MENTHU Score for the DSB need to be less than or equal to 5 bp and more than or equal to 1.50, respectively, for a positive PreMA prediction.