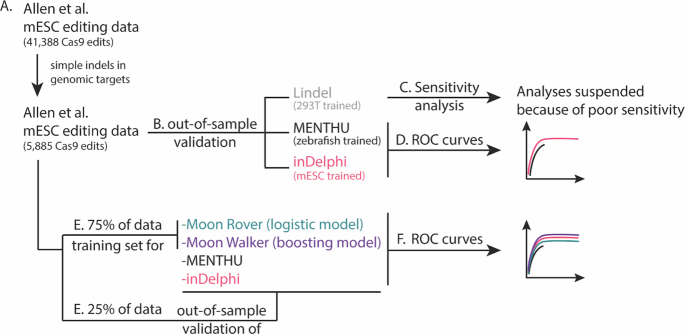
Figure 2.
Workflow of the independent assessment of the ability of MENTHU to predict PreMAs. (A) A large gene editing dataset was filtered to only include genomic DSB repair outcomes that resulted in simple indels (i.e. resulting in single deletions or insertions). (B) This dataset was used to assess the viability of MENTHU PreMA predictions in a mammalian cell system (mouse ESC cells [mESCs]), since MENTHU was originally validated in zebrafish embryos. To contextualize any MENTHU claims, the same dataset was used to generate PreMA predictions using inDelphi and Lindel, similar-purpose software tools in the recent literature. (C) Lindel predictions resulted in less than 1% sensitivity and were therefore excluded from downstream PreMA analyses. (D) Receiver Operating Characteristic (ROC) curves were used to compare the ability to predict PreMAs by MENTHU and inDelphi. (E) To investigate whether the MENTHU prediction scheme maximizes the predictive capacity of the features it uses for classification, the large dataset described in (A) was split into 75% for the training of machine learning models for PreMA predictions and 25% for the out-of-sample evaluation of these models. (F) The training set in (E) was used to train Moon Rover (a logistic regression classifier) and Moon Walker (a gradient boosting machine classifier). ROC curves for Moon Rover and Moon Walker were generated based on their predictive performance on the testing set in (E), and were plotted together with ROC curves of MENTHU and inDelphi on the same testing set for reference.