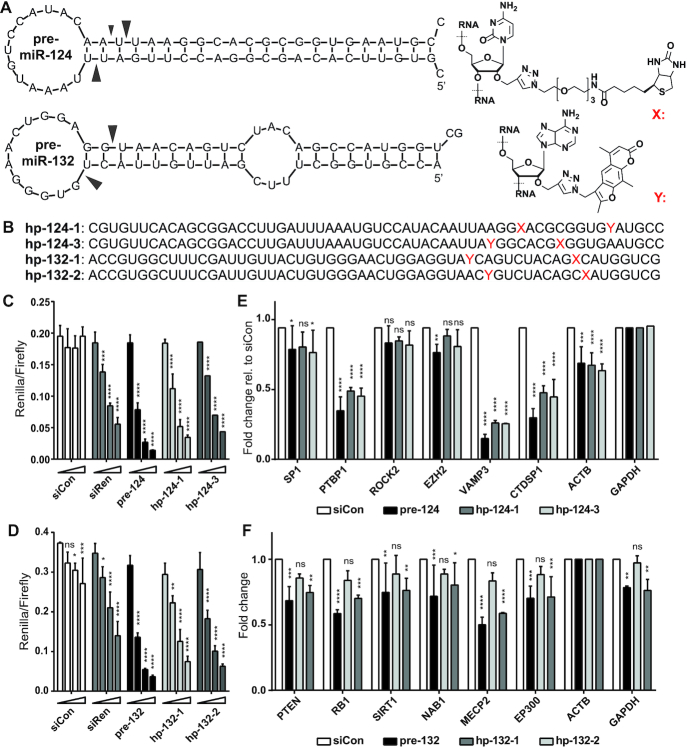
Figure 1.
Design and characterization of miR-CLIP probes for miR-124-3p and miR-132-3p. (A) Predicted structures (mFold) of pre-miR-124 and pre-miR-132 and structures of the biotin (X) and psoralen (trioxsalen)-(Y) modified bases. Main Dicer-cleavage sites in 5p- and 3p-arms according to miRBase (39) are indicated with arrows. (B) Sequences of miR-CLIP probes for miR-124-3p and miR-132-3p; cytidines labeled with biotin are indicated with X; adenosines labeled with psoralen are indicated with Y. (C andD) Luciferase reporter gene suppression by wild-type pre-miR-124 and pre-miR-132, and two miR-CLIP probes for each miRNA; HEK293T cells were co-transfected with luciferase reporter plasmids containing one reverse complementary target site against the respective miRNA and three concentrations of pre-miRNAs or miR-CLIP probes (0, 2.5, 10, 40 nM); N = 3. (E and F) Transcript levels of literature-reported miR-124 (E) and miR-132 (F) targets after transfection with 40 nM of the indicated RNA. Transcript levels were compared to transfection with negative control RNA, siCon (40); N = 3. Error bars indicate standard deviations. Asterisks denote statistical significance compared to 0 nM dose (C, D) or siCon treatment (E, F) assessed by two-way ANOVA Dunnett test whereas: ns P > 0.05, * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001.