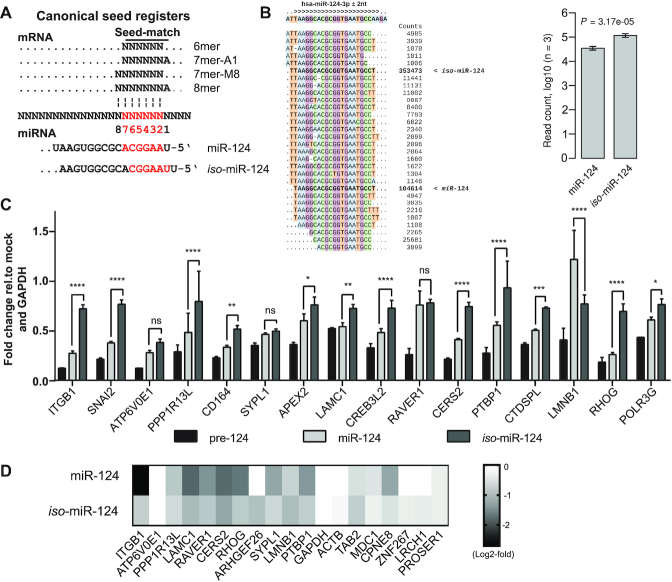
Figure 5.
Pre-miR-124 produces isomiRs with distinct targeting properties. (A) MiRNA–mRNA binding motifs via interactions involving nucleotides g1-g8 of miR-124 and iso-miR-124 (adapted from(2)). (B) Small RNA-Seq read alignments to hsa-miR-124 (left) and log10-transformed counts of small RNA-Seq reads, supporting miR-124 and iso-miR-124 in libraries extracted from HEK293T cells transfected with 40 nM pre-miR-124 (right). The indicated P value was calculated with a two-tailed, paired t-test (see Supplementary Figure S10; for clarity, nucleotides are color-coded). (C) Fold change of miR-124 target mRNAs after treating HEK293T cells with 40 nM of pre-miR124, miR-124 or iso-miR-124 duplexes. Asterisks denote significance between miR-124 and iso-miR-124 treatment. Significance calculated by two-way ANOVA Dunnett test: ns (P > 0.05), * (P ≤ 0.05), ** (P ≤ 0.01), *** (P ≤ 0.001), **** (P ≤0.0001). (D) Inhibition of selected miR-CLIP targets determined by multiplexed protein identification and quantitative analysis by tandem mass spectrometry. Heatmap represents the fold change of the designated targets in miR-124 or iso-miR-124 transfected cells with respect to mock transfected cells. Average fold change is computed from three independently transfected biological replicates of each treatment.