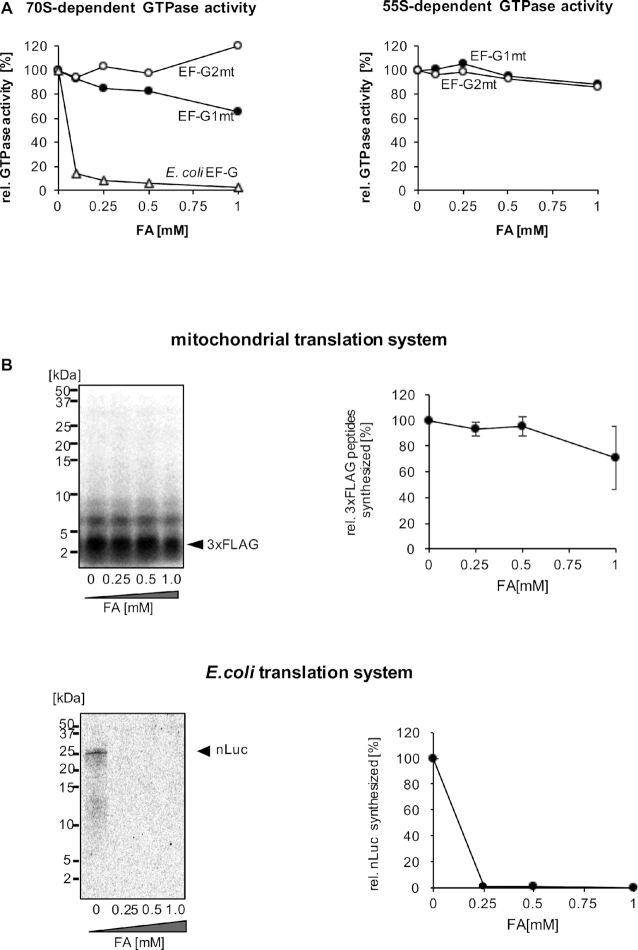
Figure 4.
Fusidic acid (FA) resistance of the reconstituted mammalian mitochondrial translation system. (A) Effect of FA on ribosome-dependent GTPase activity. E. coli EF-G/EF-G1mt/EF-G2mt (final concentration 0.5 μM) were incubated with ribosomes (final concentration 0.2 μM) and [γ-32P]GTP in the presence of the indicated amounts of FA, and the [32P]Pi release was measured. Left, assay with E. coli 70S ribosomes; right, assay with mitochondrial 55S ribosomes. The amounts of Pi released at 0 mM FA correspond to 1,400 pmol (E. coli EF-G), 650 pmol (EF-G1mt), and 170 pmol (EF-G2mt) on 70S ribosomes, and 4,300 pmol (EF-G1mt) and 1,600 pmol (EF-G2mt) on 55S ribosomes. Note that the GTPase activity of E. coli EF-G is not detectable on 55S ribosomes. (B) Effect of FA on 3xFLAG peptide synthesis (upper). 3xFLAG peptides were synthesized as in Figure 3B, in the presence of the indicated amounts of FA. After the 120 min translation reaction, the samples were treated with RNase A and resolved by Tricine SDS-PAGE (upper left), and the amounts of the synthesized 3xFLAG peptides were plotted (upper right). Error bars represent the standard deviation from three independent experiments. Effect of FA on the nLuc synthesis with E. coli translation system (PURE frex ver. 2.0) was similarly analyzed for reference (lower). The standard deviation from three independent experiments do not exceed 0.3%.