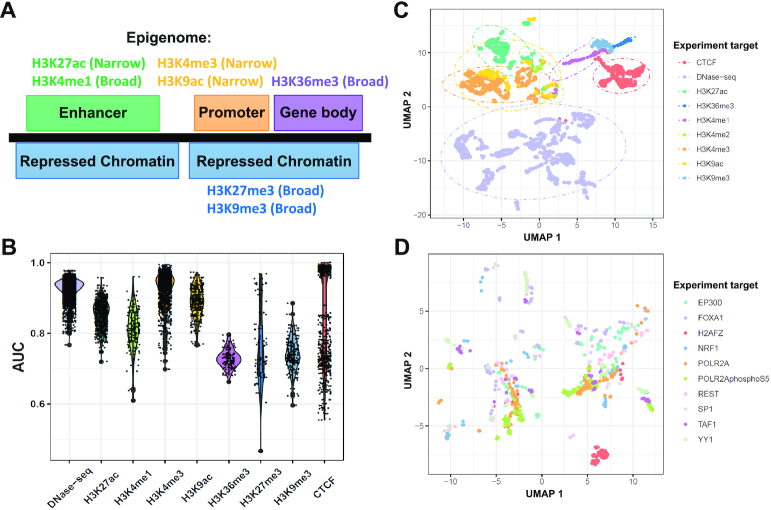
Figure 2.
Evaluation of DeepFun model across different experimental targets. (A) Overview of histone mark associated regions. (B) Evaluaton of model performance. Each violin represents the AUC value of the evaluation for a specific DNase-seq or histone mark feature. (C, D). Uniform manifold approximation and projection (UMAP) analysis of tissue and cell type specificity for the epigenomic profiles for (C) DNA accessibility and histone mark, and (D) transcription factor.