Figure 1.
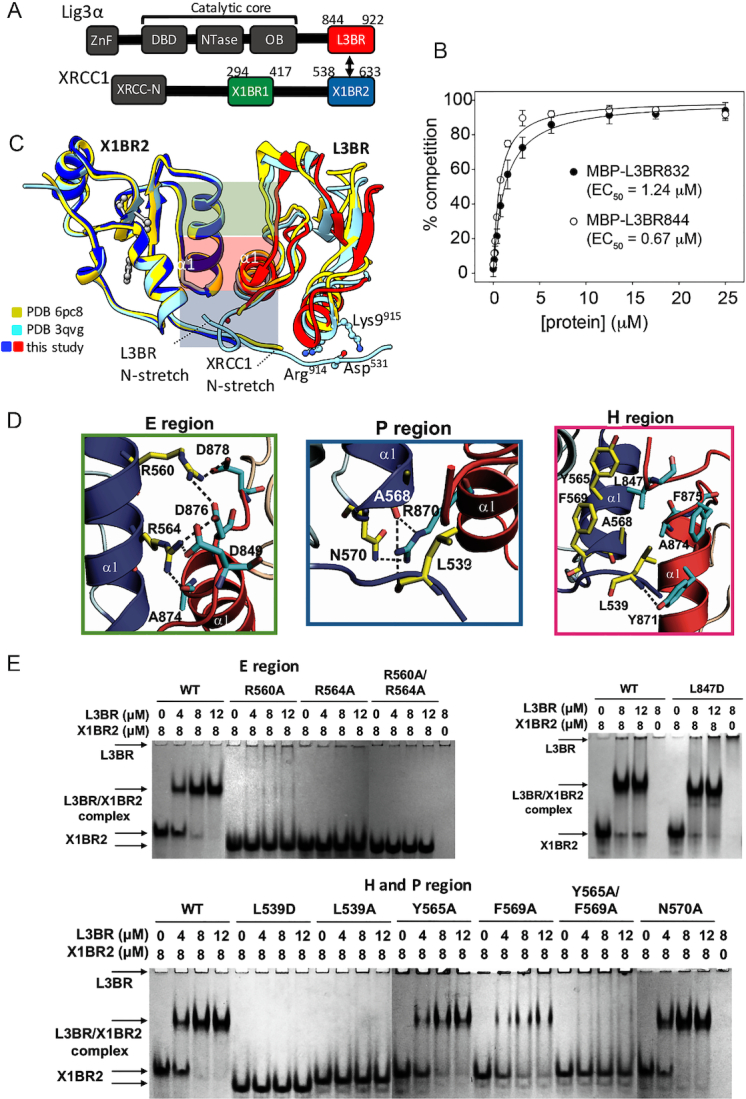
Human BRCT–BRCT heterodimer structure stably links XRCC1 and LigIIIα via a mutationally verified interface. (A) Domain organization of human XRCC1 and LigIIIα with the C-terminal BRCT domains used for crystallization (red and blue-boxes) (B) GFP-fluorescence-based competition assay (see Supplementary Figure S3) measuring specific binding affinity of L3BR832 and L3BR844 for X1BR2. The data shown represent the mean values and standard deviations from three independent experiments. (C) Structure of human XLBR-BR complex (X1BR2, blue; L3BR, red) is superimposed on to previously reported structures of heterodimers between mouse X1BR2 and human L3BR with two different lengths of N-stretch region. The different lengths of N-stretch regions of X1BR2 and L3BR constructs are highlighted. The XLBR-BR interfaces, which are classified based on the location and main type of interaction [electrostatic (E region, top, green), hydrophobic (H region, middle, purple) and polar (P region, bottom, blue) interactions], are indicated. (D) Panels show close-up of three main binding interfaces between X1BR2 and L3BR. (E) Effects of substituting residues that are located at the BRCT–BRCT interface on X1BR2–L3BR complex formation measured by native gel analysis; upper left panel, substitution of amino acids in the E region; upper right panel, substitution of Leu847 of L3BR, an equivalent of Leu539 of X1BR2; lower panel, substitution of amino acids in the H and P regions of the binding interface. The X1BR-only and L3BR-only control reactions are shown in the leftmost and rightmost lane of each gel, respectively. For residues in E region (upper left panel) or H and P regions (lower panel), gels were run with negative control (X1BR2 or L3BR alone) and positive control (with wild-type X1BR2, WT) reactions, and then combined in a single panel to compare their effects on L3BR binding. Representative gels from two independent experiments are shown. Since the theoretical pI for L3BR is 9.22 compared with 4.90 for X1BR2, L3BR does not enter the gels, which are run at pH 7.5, unless it is complexed with X1BR2.
