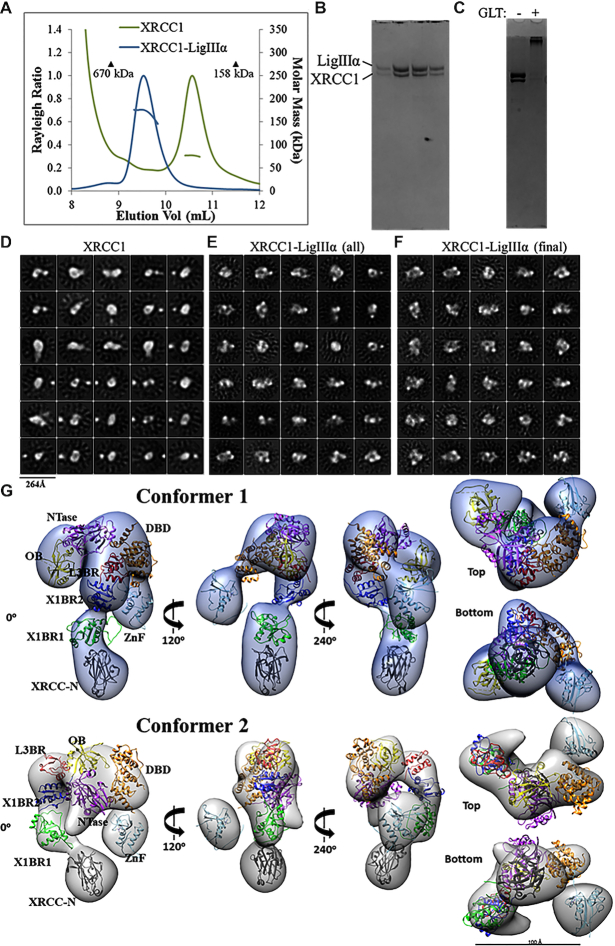
Figure 4.
Visualization of XL particles by electron microscopy reveals a dynamic domain arrangement. (A) SEC-MALS analysis shows that XRCC1 either alone or in complex with full-length LigIIIα elutes earlier than globular protein standards despite molar masses of ∼77 and ∼180 kDa, respectively. (B) The XL peak seen in (A) is composed of a heterodimer. (C) The XL dimer was chemically crosslinked prior to negative-stain EM imaging. (D) 2D classification reveals oligomeric and conformational heterogeneity in non-crosslinked XRCC1. (E) Particles belonging to non-XL 2D classes were removed from the cross-linked XL dataset. (F) The final dataset contained uniformly-sized 2D classes of particles. 3D classification and subsequent refinement resulted in two conformers of the XL complex. (G) The 3D EM maps are rotated to each display a linear protrusion from a larger area of density. The linear extension potentially represents XRCC1 domains X1BR2 (PDB ID: 3PC8 chain A), X1BR1 (PDB ID: 2D8M) and the N-terminus (PDB ID: 3K77). The LigIIIα ZnF (PDB ID: 1UW0), three domain catalytic fragment (DBD, NTase and OBD, PDB ID: 3L2P) and L3BR (PDB ID: 3PC8 chain C) domains were docked into the larger area of density of each conformer.