Figure 2.
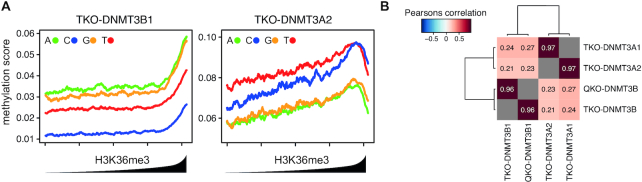
Flanking sequence preferences scale with, but are independent of the genomic location of the DNMTs. (A) Preferential de novo methylation of purines by DNMT3B is not altered by its general preference for H3K36 tri-methylated sites. Shown are de novo DNA methylation at all four CpGpN context genome-wide in relation to H3K36me3 enrichment. 1-kb-sized genomic intervals were ranked and grouped by H3K36me3 enrichment (1000 intervals per bin) and DNA methylation was calculated per bin. Lines indicate mean DNA methylation per bin in TKO cells expressing DNMT3B1 (left) or DNMT3A2 (right). (B) Heatmap indicating strong correlation in DNA sequence preference between the DNMT3A1 and DNMT3A2 isoforms, or between DNMT3B1 in presence and absence of H3K36me3. Correlations between identical samples were removed and are shown in gray.