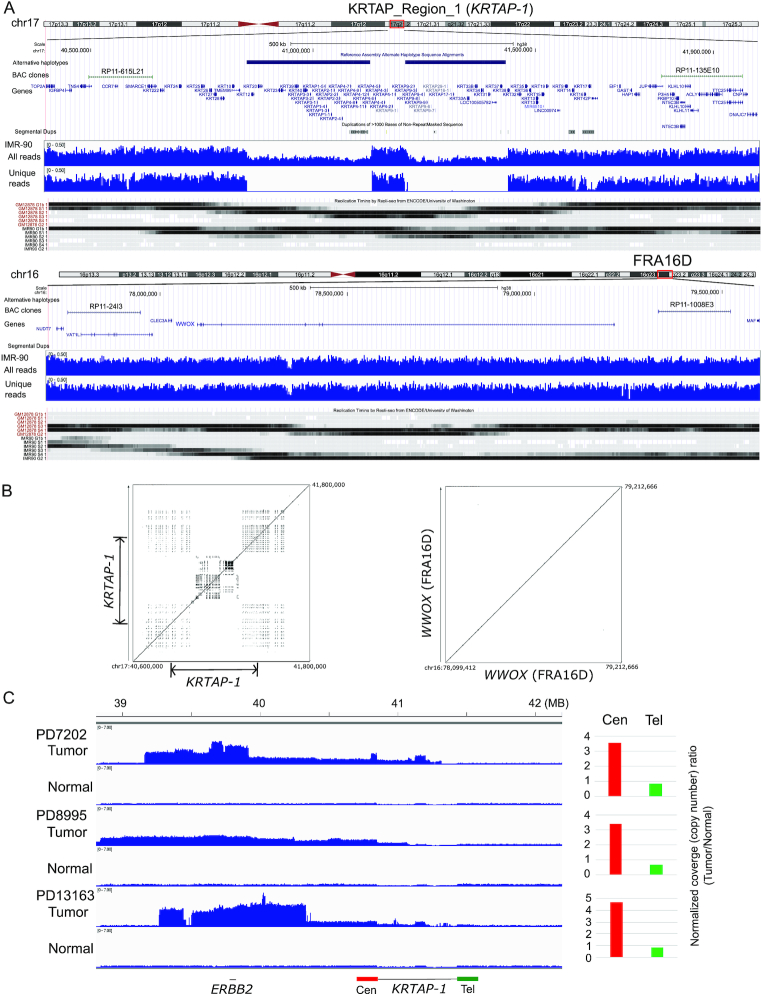
Figure 1.
KRTAP_Region_1 is a 500 kb block on chromosome 17q21.2. (A) KRTAP-1, FRA16D, and their flanking regions in hg38. (top) The locations of alternative haplotypes, BAC clones for Break-apart FISH probes, and genes within the regions are shown. (middle) Read depth of IMR-90 whole genome sequencing for total reads and for reads that are uniquely mapped (unique reads, MAPQ > 40) to the regions are shown. (bottom) The replication timings of GM12878 and IMR-90 cells from Repli-seq data (ENCODE), with the regions replicating in late G1 (top), S (four phases) and early G2 (bottom) phases are shown by shades. (B) Advanced PipMaker dot plot of KRTAP-1 and WWOX gene (FRA16D). The coordinates in the hg38 are shown. Dots represent sequences (≥100 bp without a gap) with ≥70% nucleotide identity. (C) Read coverage of the 17q12–21 region for the whole genome sequencing (WGS) data of breast tumors (European Genome-Phenome Archive data, EGAD00001001334). Normalized coverage (# of reads/kb/million reads) from three tumor/normal pairs are shown. Note that in all three tumors, the side centromeric to KRTAP-1 has greater read depth than the region telomeric to KRTAP-1. Histograms show the normalized coverage ratio between tumor DNA and normal DNA for the region centromeric to KRTAP-1 (200 kb region, Cen, in red) and telomeric to KRTAP-1 (Tel, in green).