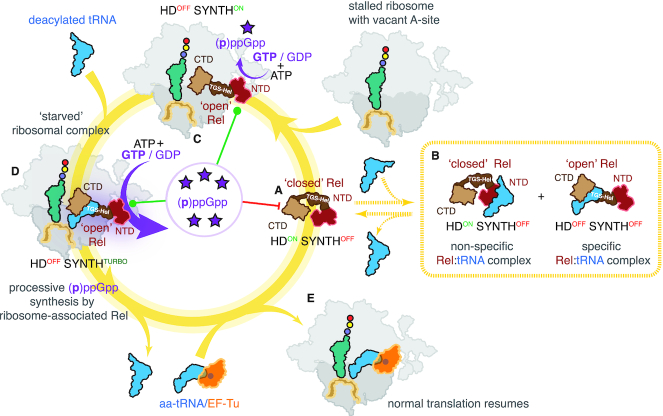
Figure 6.
Model of Rel regulation by starved ribosomal complexes. (A) Off the ribosome Rel assumes a ‘closed’ conformation concealing the tRNA-binding TGS and Helical domains. The factor is in hydrolytically active (HDON SYNTHOFF). (B) ‘Closed’ Rel can bind deacylated tRNA off the ribosome which suppresses the HD activity (HDOFF SYNTHOFF) as well as non-specifically bind tRNA through the NTD region. Dashed lines signify that while the interactions are observed in biochemical assays, their physiological relevance is unclear. (C) Upon binding to the vacant A-site of a starved ribosome, Rel switches to an ‘open’ conformation which stimulates the (p)ppGpp synthesis and inhibits (p)ppGpp hydrolysis (HDOFF SYNTHON). (D) Opening up primes Rel for specific recognition of deacylated tRNA by its TGS and Helical domains. Binding of deacylated tRNA stabilizes Rel on the ribosome and leads to full activation of the processive (p)ppGpp synthesis activity (HDOFF SYNTHTURBO). (E) Translation resumes once the aminoacylation levels are restored. Accumulating pppGpp acts as an allosteric regulator of Rel. In the case of the HDOFF factor bound to either vacant ribosomal A-site associated with starved complexes, the alarmone binds to the allosteric site in the NTD domain region to stimulate (p)ppGpp synthesis activity. In the case of the HDON free factor, the alarmone acts as an HD substrate, thus suppressing the synthesis activity via inter-NTD regulation.