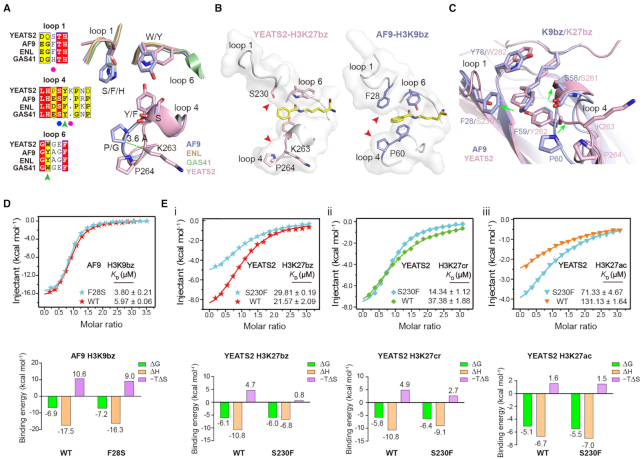
Figure 5.
A ‘tip-sensor’ mechanism determines acylation type selectivity of YEATS domains. (A) Sequence and structural alignments of reader pocket loops among four human YEATS domains. Magenta circles, ‘tip-sensor’ residues; Blue hexagon, conserved Ser residue for acyllysine amide recognition; Green arrowhead, aromatic sandwiching residues; Light blue, AF9YEATS; Wheat, ENLYEATS (PDB ID: 5J9S); Pale green, GAS41YEATS (PDB: 5XTZ); Light pink, YEATS2YEATS. (B) Comparison of reader pocket between YEATS2YEATS and AF9YEATS. Red arrowheads indicate a wide and narrow end-opening of YEATS2 and AF9, respectively. (C) Structure alignment of YEATS2YEATS-H3K27bz with AF9YEATS-H3K9bz centred on the recognition pocket. Green arrows indicate conformational variations between the two proteins. (D) ITC assays of Kbz binding by WT AF9YEATS and its F28S mutant. Mean KD and standard deviation are shown (N = 4). Thermodynamic parameters of each titration are illustrated below. (E) ITC assays of (i) Kbz, (ii) Kcr and (iii) Kac binding by WT YEATS2YEATS and its S230F mutant. Mean KD and standard deviation are shown (N ≥ 2). Thermodynamic parameters of each titration are illustrated below.