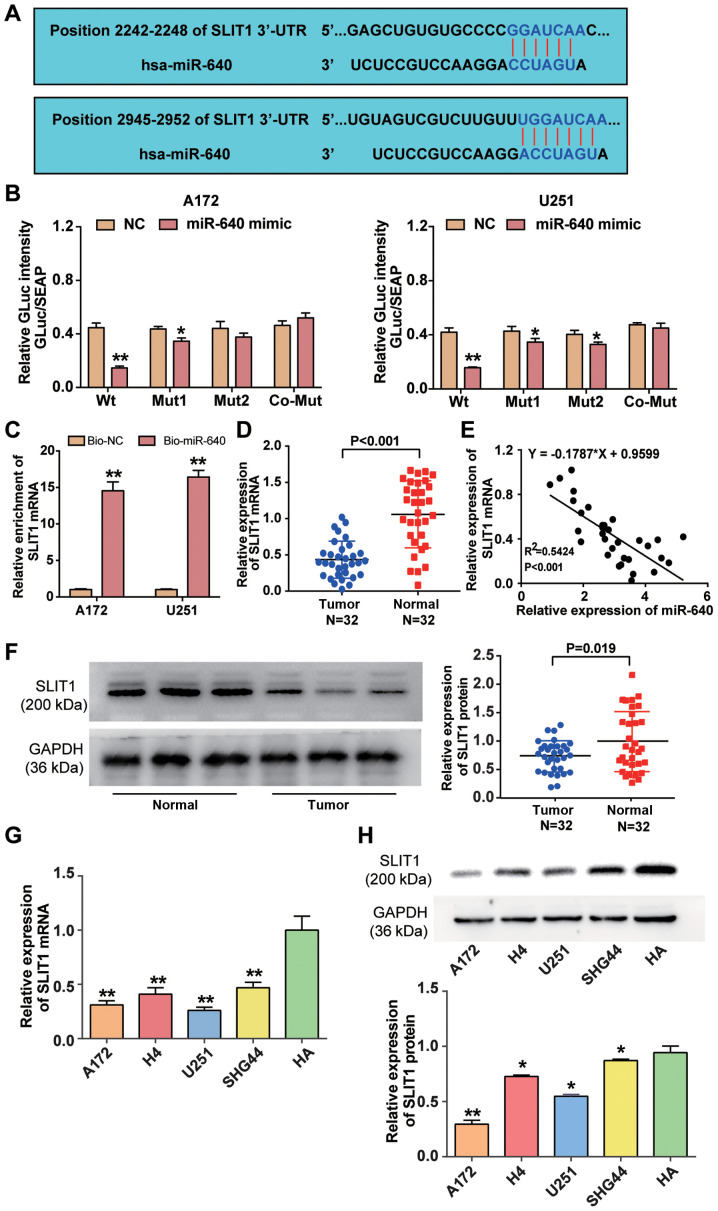
Figure 3.
miR-640 directly binds to SLIT1 3′-UTR. (A) The potential binding site between miR-640 and SLIT1 was predicted using miRDB. (B) The potential binding site between miR-640 and 3′-UTR of SLIT1 was determined via the Dual-luciferase reporter assay. (C) The RNA pull-down assay was performed to detect SLIT1 enrichment in A172 and U251 cells. (D) RT-qPCR analysis was performed to detect SLIT1 expression in glioblastoma tissues and normal tissues. (E) Spearman's correlation analysis was performed to determine the association between miR-640 and SLIT1 expression levels. (F) Western blot analysis was performed to detect SLIT1 protein expression in glioblastoma tissues and normal tissues. (G) RT-qPCR analysis was performed to detect SLIT1 mRNA expression in the glioblastoma cell lines (A172, H4, U251 and SHG44) and normal astrocyte HA cells. **P<0.001 vs. HA. (H) The expression level of SLIT1 protein in glioblastoma cell lines (A172, H4, U251 and SHG44) and normal astrocyte cell line HA was measured using a western blot system. Data are presented as the mean ± standard deviation (n=3). *P<0.05 vs. co-transfection with Wt and miR-640 mimic or HA; **P<0.001 vs. co-transfection with Wt and miR-640 mimic, Bio-NC or HA. miR, microRNA; SLIT1, Slit guidance ligand 1; UTR, untranslated region; Wt, wild-type; Mut, mutant; Bio-NC, biotin-coupled negative control; RT-qPCR, reverse transcription-quantitative PCR.