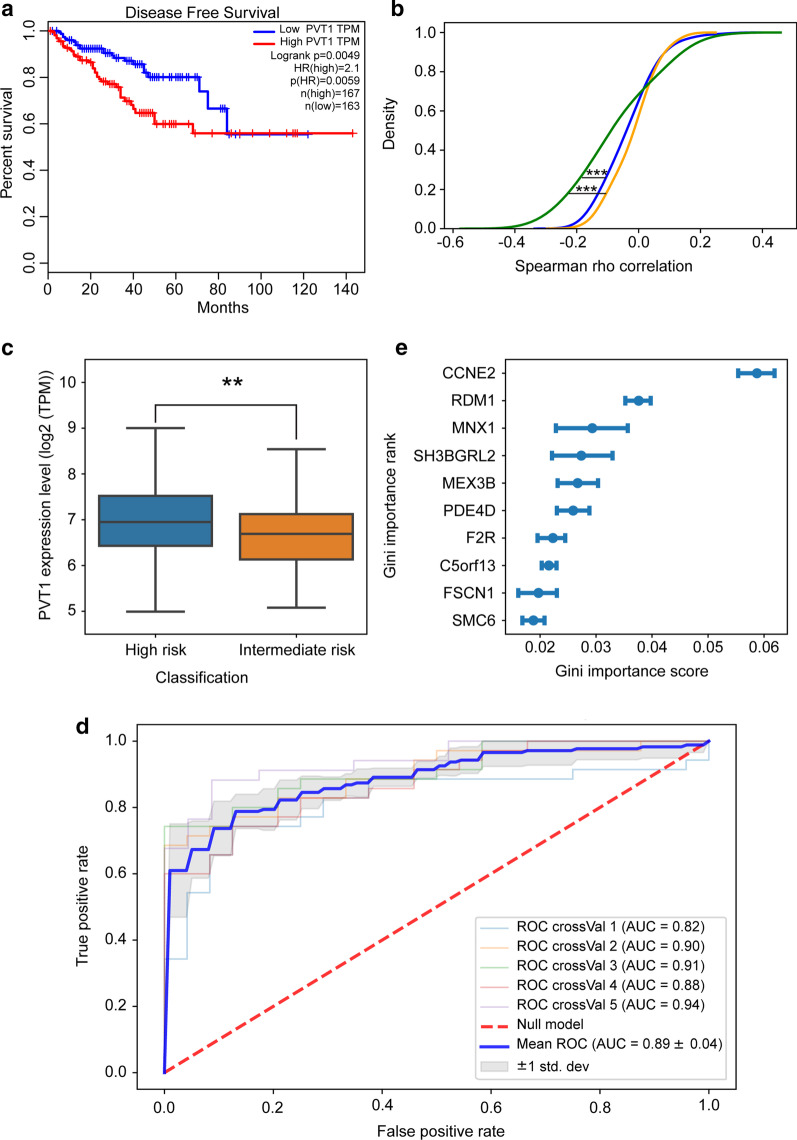
Fig. 4.
Genes de-repressed by PVT1 knockdown in LNCaP cells had their expression in the TCGA prostate adenocarcinoma dataset correlated to tumor risk. a Kaplan–Meier disease-free survival analysis for TCGA-PRAD patients based on the PVT1 expression level in the tumors of the first 3-quantile low-PVT1 samples (blue, n = 163) and third 3-quantile high-PVT1 samples (red, n = 167). b Cumulative density distribution of Spearman rho correlation between the expression levels of PVT1 and each of the 121 genes upregulated after PVT1 depletion in LNCaP cells. Correlation was calculated for each gene in the 121-gene-set across all TCGA-PRAD tumors from patients separated into three groups comprised of samples with low-, medium- or high-PVT1 expression (orange, blue and green lines, respectively). The Kolmogorov–Smirnov test (two-tail ks-test) was applied to calculate the significance of the difference between the groups’ distribution (orange***, P value < 4.5 × 10–6; blue***, P value < 9.0 × 10–4). c PVT1 expression level in TCGA-PRAD tumor samples with high (n = 174) and intermediate risk (n = 119). Significance was calculated using a two-tail t-test, **P < 0.01. d Classification of intermediate- and high-risk TCGA-PRAD tumors using a Random Forest machine-learning model. The ROC curves show the area under the curve (AUC) for the Random Forest model classification performance in a 5-way cross-validation (crossVal) scheme. Expression of the 121-gene-set across all intermediate- and high-risk TCGA-PRAD tumors were used as the input features for the machine-learning model. The 5-way cross-validation mean is indicated by the blue line, and the gray shade represents the validation standard deviation. e Rank of the top 10 most predictive genes in the Random Forest tumor classification analysis. Each blue point and error bars represent the mean ± S.D. of the Gini importance score for that gene, obtained in the 5-way cross-validation scheme