Fig. 7. Quantification of lipid mass in PITPα+/+ and PITPα−/− brain and liver.
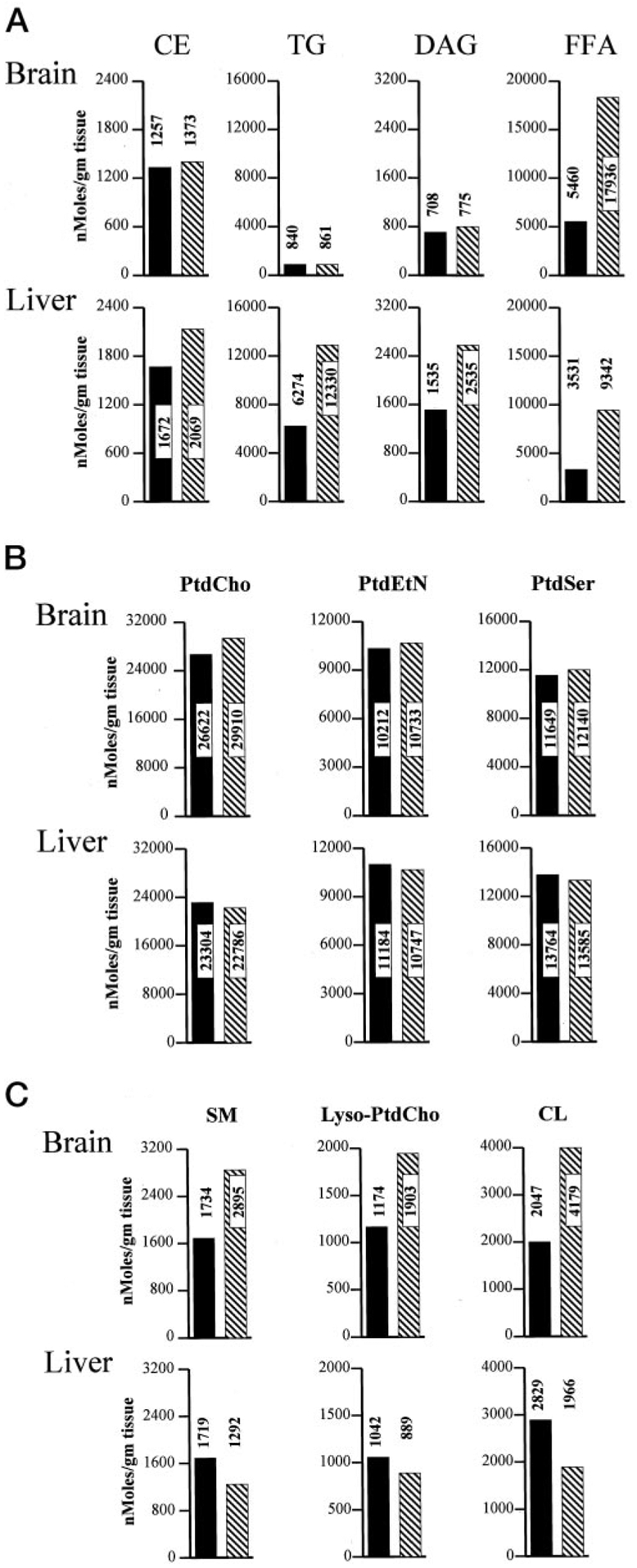
Brains and livers were collected from each of four freshly euthanized non-fasted PITPα+/+ and PITPα−/− mice and combined to generate defined tissue pools for each genotype. Pools were frozen and quantified for individual lipid species and FFAs via the Lipomics Technologies, Inc., TrueMass™ protocol. Individual lipid species are given at the top, and brain and liver values for each lipid species are given in a vertical column below each heading. Brain values are represented in the top panels, and the liver values are in the bottom panels as indicated. All lipid species are quantified as nanomoles per g of tissue. PITPα+/+ and PITPα−/− values are indicated by solid bars and hatched bars, respectively. Numerical values for each bar are given. A, neutral lipid mass measurements. B, polar lipids whose mass is unaltered in PITPα+/+ versus PITPα−/− tissues. C, polar lipids whose mass is altered in PITPα+/+ versus PITPα−/− tissues. SM, sphingomyelin; PtdEtn, phosphatidylethanolamine; PtdSer, phosphatidylserine.
