Abstract
SARS-CoV-2 epidemics quickly propagated worldwide, sorting virus genomic variants in newly established propagules of infections. Stochasticity in transmission within and between countries or an actual selective advantage could explain the global high frequency reached by some genomic variants. Using statistical analyses, demographic reconstructions, and molecular dynamics simulations, we show that the globally invasive G614 spike variant 1) underwent a significant demographic expansion in most countries explained neither by stochastic effects nor by overrepresentation in clinical samples, 2) increases the spike S1/S2 furin-like site conformational plasticity (short-range effect), and 3) modifies the internal motion of the receptor-binding domain affecting its cross-connection with other functional domains (long-range effect). Our results support the hypothesis of a selective advantage at the basis of the spread of the G614 variant, which we suggest may be due to structural modification of the spike protein at the S1/S2 proteolytic site, and provide structural information to guide the design of variant-specific drugs.
Keywords: SARS-Cov-2 evolution, population dynamics, generalized linear mixed models, coalescent-based inference, molecular dynamics
Introduction
After appearing in Wuhan, China, in late 2019, SARS-CoV-2 (ZhOu et al. 2020), a highly contagious (D’Arienzo and Coniglio 2020) coronavirus (CoV) causing severe acute respiratory syndrome (COVID-19), spread worldwide and rapidly emerged as a dramatic pandemic, officially acknowledged on March 11, 2020 (World Health Organization [WHO] 2020).
All CoVs present a spike glycoprotein on their surface. The spike is a trimer, whose monomers are composed of two subunits (S1 and S2) each. Only one of the monomers shows the receptor-binding domain (RBD) in the activated configuration (up; fig. 1A) (Berry et al. 2004; Pak et al. 2009; Walls et al. 2017; Song et al. 2018; Wrapp et al. 2020), necessary to contact the human angiotensin-converting enzyme 2 (ACE2) (Walls et al. 2020; ZhOu et al. 2020) and mediate host cell invasion. Spike proteins must be primed before they can be triggered to induce fusion between the viral and the host cellular membranes (Simmons et al. 2013; Hoffmann, Kleine-Weber, Schroeder, et al. 2020). Priming involves a proteolytic event at a cleavage site (S2′), mediated by the cellular serine protease TMPRSS2 (White and Whittaker 2016). A novel furin-like cleavage site (S1/S2), an exposed loop harboring multiple arginine residues (PRRAR/S) (fig. 1B and C) (Walls et al. 2020; Wrapp et al. 2020), has been suggested to be a key element enhancing SARS-CoV-2 infectivity (Andersen et al. 2020; Coutard et al. 2020; Peacock et al. 2020). This cleavage site has been observed, though with different amino acid sequences, in distantly related CoVs, like MERS-CoV and HKU1-CoV, but not in those viruses that are the most closely related to SARS-CoV-2 (Andersen et al. 2020; Coutard et al. 2020; Xiao et al. 2020). Thus, the SARS-CoV-2 spike appears to be activated by a two-step process: a priming cleavage by furin-like protease at the S1/S2 site and an activating TMPRSS2-mediated cleavage at the S2′ site during membrane fusion (Hoffmann, Kleine-Weber, and Pöhlmann 2020; Ou et al. 2020).
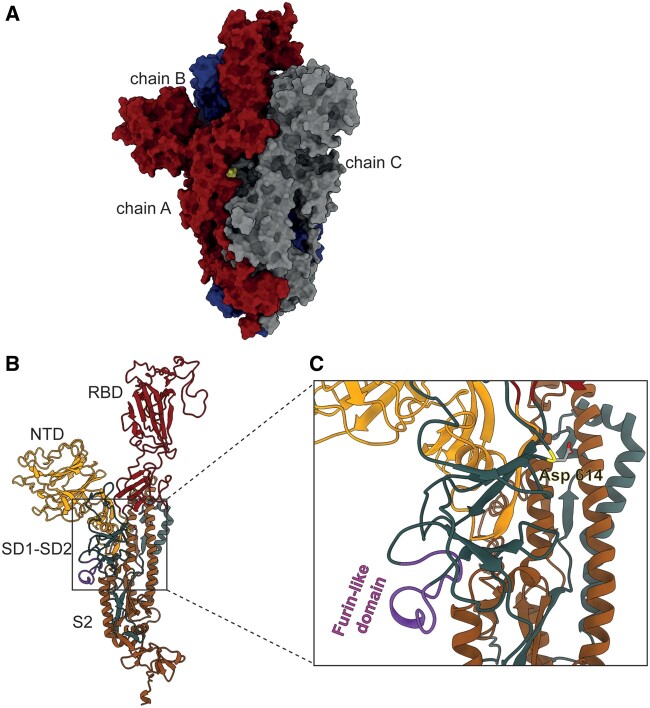
Fig. 1.
3D structure of the spike protein. (A) Molecular surface of the SARS-CoV-2 spike protein. Chain A (red) is in the up conformation, whereas chains B (blue) and C (gray) are in the down conformation. Location of D614G substitution is shown (yellow). (B) Domain subdivision of the S1 and S2 subunits of the monomer that upon cleavage by host proteases remain noncovalently bonded. The S1 subunit contains an NTD (dark yellow), a RBD (dark red) that drives host cell tropism and a C-terminal domain further subdivided into domains SD1 and SD2 (petrol green), harboring the furin-like domain (purple). The S2 subunit mainly consists of heptad repeat (HR) regions involved in membrane fusion (Liu et al. 2004; Belouzard et al. 2012; Li 2016), of which only the HR1 was resolved (orange). Monomers can exist in two main metastable conformations: down, with the RBD tightly packed against the NTD, and up representing the active form corresponding to the receptor-accessible configuration (Berry et al. 2004; Pak et al. 2009; Walls et al. 2017; Song et al. 2018; Wrapp et al. 2020). (C) Close-up on the region where the D614G variant (Asp 614; yellow) is located. The furin-like domain is highlighted (purple).
A novel variant of the SARS-CoV-2 spike protein was first observed in Bavaria, Germany (EPI_ISL_406862) and Shanghai, China (EPI_ISL_422425, 416327, 416334) in late January 2020 (GISAID EpiFluTM Database; www.gisaid.org, last accessed on 27/09/2020). A few weeks later, the viral clade carrying this variant emerged as the most abundant in Europe (Becerra-Flores and Cardozo 2020; Brufsky 2020; Chiara et al. 2020; Pachetti et al. 2020), and in April it was acknowledged as the prevailing one worldwide(Becerra-Flores and Cardozo 2020; Brufsky 2020; Chiara et al. 2020; Korber et al. 2020; Pachetti et al. 2020). This SARS-CoV-2 clade is characterized by a nonsynonymous nucleotide transition A → G at the genomic position 23403 (Wuhan reference genome) (Wu et al. 2020), replacing an aspartic acid at the spike position 614 (fig. 1) with a glycine (hereafter, we refer to the novel spike protein variant as G614, whereas the ancestral state is indicated as D614). As G614 prevails in every region where it seeded the epidemic and also where it was introduced after the alternative D614, a selective advantage for this novel variant, and/or its linkage group (Pachetti et al. 2020), has been suggested (Korber et al. 2020; VasilarOu et al. 2020). However, stochastic processes, such as sequential founder effect (Chiara et al. 2020) or shared genetic drift due to abundant gene flow among geographic regions, have not been explicitly tested as alternative causes for the observed global frequency increase of G614. Most importantly, a functional explanation for the invasiveness of G614 is still missing.
Here, we initially demonstrate that the global increase of G614 variant in the genomic samples can be explained neither by a founder effect or shared drift due to high connectivity among epidemics in different geographic areas nor by overrepresentation in clinical samples.
Then, we show that the D → G mutation at residue 614 has both short- and long-range effects on the dynamics of the spike protein, affecting the S1/S2 furin-like cleavage site and the RBD spatial conformation, respectively. In the G614 variant, the multibasic furin-like domain is much more exposed than in the D614 one, and the position of the cleavage residue R685 is strongly stabilized by the electrostatic interaction with residue D663, likely facilitating the recognition by the protease. In addition, our results support the hypothesis that the glycine in position 614 produces a more open conformation in the activated RBD, though not resulting in a higher binding affinity of the spike for the ACE2 receptor, as shown by our molecular docking analysis.
Results and Discussion
The Global Increase of G614 Frequency Is Not Due to Random Genetic Drift
To test whether selection might have played a role in the frequency increase of G614, we took advantage of the fact that different areas of the world provided different natural sample populations, albeit nonindependent ones. Thus, we aimed at assessing whether a global increase of G614 could be detected even when accounting for the nonindependence of the epidemiological dynamics between different regions. Specifically, we tested whether the frequency increase of the G614 variant can be explained by random genetic drift and connectivity among local epidemics by fitting generalized linear mixed models (GLMM) with binomial error structure on a sample of 68,491 viral genomic sequences from 76 geographic areas (countries, US states, or Chinese provinces, see Materials and Methods). We modeled the probability that a given sample presented the G614 variant (i.e., the relative frequency of the G614 allele) as a function of time (days). Genetic drift within each geographic area was modeled as a random slope term for each geographic area through time, so that the model allows the frequency of G614 within each area to have its own temporal trend. To capture the effect of shared drift among geographic areas (i.e., the level of nonindependence between the epidemics in different areas due to gene flow) we modeled the covariance of DNA sequence variation between geographic areas with different metrics, resulting in different covariance matrices (supplementary figs. S1–S6, Supplementary Material online). For all these model designs, a significantly positive logistic slope of the global frequency of G614 over time, even when stochastic differences between geographic areas and their connectivity are accounted for, indicates that the modeled stochastic effects are not sufficient to explain the observed increase in frequency. First, we analyzed our full data set implementing, as a predictor, a covariance matrix (Pickrell and Pritchard 2012) (see Materials and Methods) based on the sample allele frequency in viral genomes from different geographic areas (Cov (all) in fig. 2A). We found that the change in frequency of the G614 allele cannot be explained by random drift or connectivity alone (GLMM: logistic slope per day = 0.06, SE = 0.01, 95% CI = 0.03–0.08). Plotting the predicted effects for each geographic sample highlights how all samples show a clear increase in the frequency of the G allele, although the fitted slope varies among them (fig. 2B). The only exception is Iceland, where a decreasing trend over the narrow sampled time window (February 27–March 29, 2020) can be observed. Assuming that natural selection is the sole driver of allele frequency change (after controlling for stochastic effects), we note that our logistic slope translates into an estimate of the selection coefficient (s) when appropriate values for the basic reproduction number and serial interval are considered (Volz et al. 2020). Assuming a basic reproduction number between 2.0 and 3.5 and a serial interval of 6.5 days (Flaxman et al. 2020; Volz et al. 2020), our global mean estimate corresponds to s in between 0.31 and 0.55 (0.24–0.67 when the 95% confidence limits are considered; as a comparison, Volz et al. [2020] estimated s to be between 0.06 and 0.56 in the United Kingdom; see supplementary fig. S8, Supplementary Material online, for estimates of s for each geographic area). As shown in figure 2A, our results also held (i.e., logistic slope per day has positive values) when 1) we computed covariances or genetic distances between populations using G614 sequences only (Cov (G)); 2) we restricted the analyses to sequence data collected in April 2020, when most countries applied very restrictive travel limitations, thus reducing gene flow (Cov (G, Apr)); 3) different metrics were used to account for gene flow among geographic areas (Fst (all), Fst (G), Fst (G, Apr); see Materials and Methods for details); and 4) geographic distances were used as proxy of gene flow among areas (Geo, Geo (Apr)). In addition, we also modeled nonindependence between countries as a single source of G614 contributing viral genomes to the different countries (source–sink model; see Materials and Methods for details), testing for three different potential source countries selected for their high estimated G614 frequency at the beginning of the pandemic spread (Belgium and Iceland) or high number of cases reported early in the pandemic (Italy). None of these source–sink models (Iceland: Akaike information criterion [AIC] = 40,949, Bayesian information criterion [BIC] = 40,995; Belgium: AIC = 41,005, BIC = 41,050; Italy: AIC = 41,017, BIC = 41,063) improved the fit over a model in which time itself was the fixed effect predictor (AIC = 40,764, BIC = 40,809), indicating that founder effect followed by global spread from a single source does not explain the steady worldwide increase in G614. All in all, our results show that the observed increase in frequency of G614 across different geographic areas cannot be explained by any of the metrics we implemented to account for gene flow among local epidemics, hence supporting the hypothesis of a selective advantage for this variant. We caution that extreme epidemiological patterns (e.g., a single geographic area asymmetrically spreading G614 migrants worldwide associated with strong discontinuities in the timing of gene flow) may have escaped control by our model design. However, as significant global increase of the G614 variant was also observed in the April 2020 subset (i.e., during worldwide travel ban) and that our source–sink model (i.e., one country contributing the most to the rest of the world epidemics) was not favored, such unmodeled epidemiological features are unlikely to explain our results.
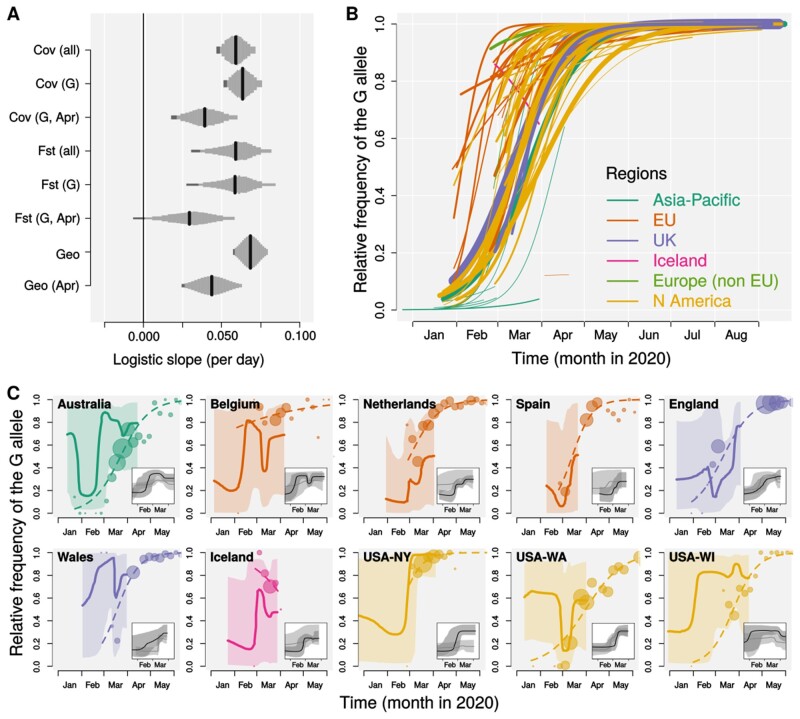
Fig. 2.
Frequency increase of the spike G614. (A) Estimated logistic slopes for the relative frequency of the G614 variant over time from mixed models. Shared drift was modeled by covariance matrix (Cov), FST, or geographic distance (Geo); genetic differentiation metrics (FST or Cov) were computed on the full sequence data set (all) or sequences with the G614 allele only (G); (G, Apr) indicates that the differentiation metrics were computed on G614 sequences only and that the model was fitted on a subset of sequences collected in April 2020. Black bars indicate mean estimates and gray bars show Bayesian estimates of posterior distributions (or Gaussian distribution of estimate based on SE for the Geo models) comprised between 2.5% and 97.5% quantiles (95% CI). Dark gray areas highlight portions of the posterior distribution between 2.5% and 5%. Note that, for all models, >95% of the posterior distribution is above 0. (B) Fitted logistic growth for each of 76 geographic areas estimated by the Cov (all) pGLMM; line width is proportional to square root of sample size; line colors indicate different geographic areas as show in legend. (C) Comparison of the frequency of the G614 variant in the genomic samples (circles: raw data aggregated in 5-day intervals, size proportional to square root of sample size; dashed lines: pGLMM fit as in panel B) and in the population as BSP ratio (solid lines: medians with line width proportional to square root of sample size; shaded areas: 99.75% confidence intervals; see Materials and Methods for details) for ten areas with at least 30 sequenced genomes per variant. Demographic reconstructions, as inferred by Bayesian Skyline plot for the D614 (gray) and G614 (black), median (solid line) and 95% CI (shaded area), are shown as insets within each area’s panel (see also supplementary fig. S10, Supplementary Material online).
Among 21 nucleotide polymorphisms with minor allele frequency >0.05 in our data set, we found that the G614D polymorphism and those in the same linkage group (genomic positions 241, 3037, and 14408) (Korber et al. 2020; Pachetti et al. 2020; Zhu et al. 2020) show, by far, the clearest signal of frequency change across the 76 investigated geographic areas (supplementary fig. S9, Supplementary Material online). Although any of the linked changes could contribute to the observed frequency increase of the whole linkage group, only the C14408T is also a nonsynonymous mutation in the nsp12 gene coding for the RNA-dependent RNA polymerase. The C241T occurs in the short noncoding region before the ORF1a and C3037T is a synonymous change within ORF1a. Pachetti et al. (2020) suggested an effect of the C14408T on virus replication efficiency or fidelity, but their results, based on the number of mutations co-occurring with the C14408T, are influenced by the early appearance of the whole linkage group upon European colonization. On the other hand, being responsible of the first contact between the virion and the host cell (Walls et al. 2020), the spike protein is the first candidate to investigate for functional modifications in the early phase of an epidemic when the novel host infection mechanism is being refined as was also suggested for the SARS-CoV epidemic (He et al. 2004).
The G614 Frequency Increase in Genomic Samples Is Not Due to Sampling Bias
The G614 fitted logistic growth in each genomic sample is largely consistent with its relative dynamics in the respective population, as reconstructed by a coalescent-based approach (Bayesian Skyline plots—BSP) (Drummond et al. 2005) (fig. 2C and supplementary fig. S10, Supplementary Material online). This result argues against a sampling bias related to G614 symptom severity. In fact, as genomic samples available through the GISAID EpiFlu Database are mostly from hospitalized patients or patients showing COVID-19 symptoms, the apparent increase in frequency of the G614 variant could have resulted from more severe COVID-19 outcomes associated with G614, which would increase the prevalence of this variant in samples from patients seeking treatment. Our result extends to a global scale the lack of association between G614 and COVID-19 symptoms severity observed in UK hospitals (Korber et al. 2020; Volz et al. 2020), indirectly excluding (i.e., as no analysis on actual pathogenicity the two spike variants were performed in our study) this potential sampling bias as a driver of the observed G614 frequency increase in the genomic samples. In Iceland and the Netherlands only, the BSP reconstructions show a lower prevalence of the G variant in the population than in the genomic samples (fig. 2C and supplementary fig. S10, Supplementary Material online). We note that Iceland is the only area showing a clear decline in the relative frequency of G614 in the sample, which is also tracked by the BSP, but the case of Netherlands (one of the countries contributing the most to virus sequencing from the very beginning of the outbreak; GISAID Database) is more difficult to explain. Although our results are supported by direct observations (Volz et al. 2020), we note that the Bayesian approach we implemented here is not accounting for multiple coalescences or population substructure. In addition, the large uncertainty in our demographic reconstructions, which is inherent to any coalescent-based inference, has been exacerbated by our highly conservative 99.75% confidence intervals calculation. While carefully considering these aspects, we suggest that the frequency increase of G614 in COVID-19 genomic samples is not driven by an overrepresentation in clinical samples due to higher symptom severity.
The Short-Range Effect of G614 Makes the Furin-like Cleavage Site More Accessible
As shown by 0.5 µs of molecular dynamics (MD) simulations for each of the G614 and D614 variants, the substitution of an aspartic acid (D) with a glycine (G) at position 614 modifies the whole fluctuation profile of the spike trimers (supplementary fig. S11A, Supplementary Material online), especially in the region downstream of the mutation (residues 622–642): In chain A, where the RBD is in the up conformation, this amino acid substitution increases the fluctuations, whereas in chains B and C where the RBD is not activated, it, instead, reduces them (supplementary fig. S11B, Supplementary Material online). The altered profile of the fluctuation extends farther downstream (residues 675–692; supplementary fig. S11B, Supplementary Material online), affecting the furin-like domain. Furthermore, an increased mobility of the furin-like domain is tracked only in RBD-activated chain A, reflecting the ability of G614 to affect the loop in the active chain, thus increasing its functional efficiency.
The increased flexibility in residues 675–692 makes the furin-like domain more solvent exposed, with an increase of the accessible surface (SAS) area of approximately 1.0 ± 0.7 nm2 in G614 as compared with D614 (fig. 3A and supplementary movie M1, Supplementary Material online). In D614, this domain is tightly bound to the protein surface, whereas in G614 the loop samples a larger conformational space. In the latter, the loop can detach from the protein and protrude toward the solvent, thus increasing its accessibility (fig. 3B). This broader movement can be appreciated by plotting the distance between residue S680, positioned at the tip of the loop and flanking the furin-like domain, and S940, located on the heptad-repeat 1 domain (HR1): This distance significantly increases in the G614 variant reaching a mean value of 2.1 ± 3 nm (fig. 3C and D).
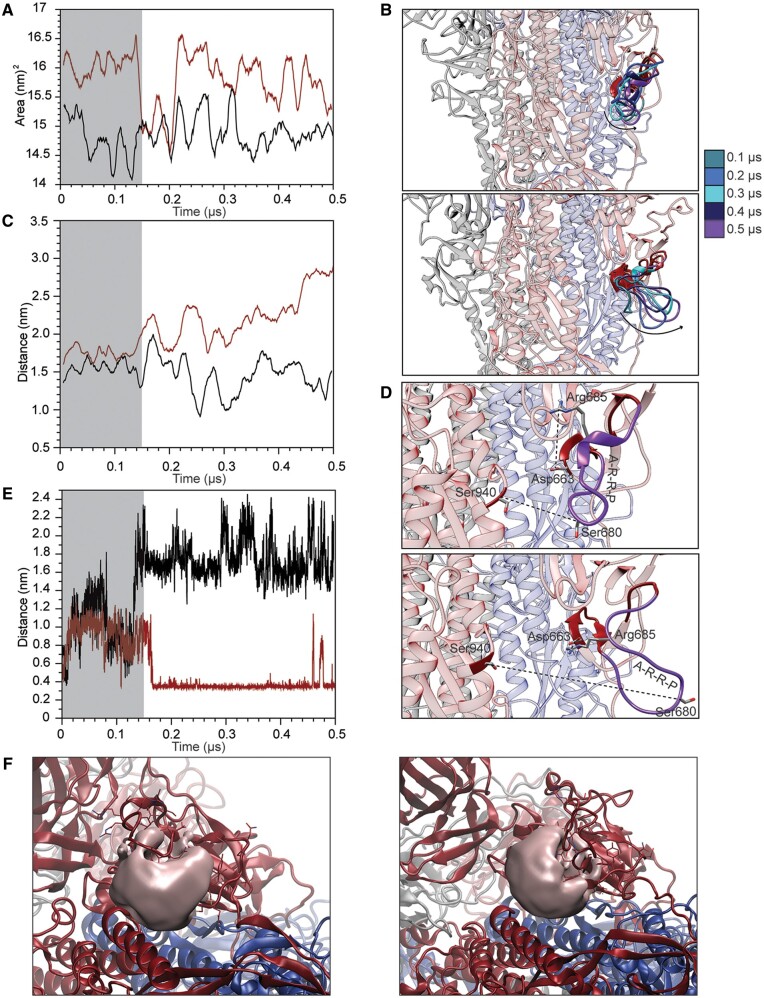
Fig. 3.
Analysis of the furin-like domain. (A) Variation of the solvent accessible surface area of residues 675–692 in the D614 (black) and G614 (red) spike protein variants. (B) Superposition of the structures extracted from the D614 (upper panel) and G614 (lower panel) trajectories showing the displacement of the furin-like domain. (C) Variation of the atomic distance calculated as a function of time between the lateral chains of S680 and S940. (D) Representative snapshots showing the relative positions of residues S680–S940 and R685–D663 in structures extracted from the D614 (upper panel) and G614 (lower panel) trajectories. (E) Variation of the atomic distance calculated as a function of time between the lateral chains of R685 and D663. (F) Representative snapshot showing the volume of the cavity formed between the furin-like domain and the surrounding structural elements in D614 (left panel) and G614 (right panel).
In addition, as a consequence of this displacement of the loop, R685, the residue of the furin-like domain directly targeted by the protease, is fixed by an electrostatic interaction formed with D663 (fig. 3D). This salt bridge is present in the G614 variant and detected for 70% of the time, whereas it is never observed in the D614 variant (fig. 3E and supplementary movie M1, Supplementary Material online). Finally, we detect an increase in the volume of the cavity formed by the multibasic loop and the surrounding structural elements. At 0.5 µs of the simulation (i.e., at the end of the simulation), we measured a volume of 1.46 and 1.7 nm3 for the D614 and G614 variants, respectively (fig. 3F). The opening of the loop in the furin-like domain increases the size of the channel at the interface with the HR1 domain where the protease can be accommodated.
Taken together, these data suggest that the presence of a glycine in position 614 has a direct effect on the dynamics of the furin-like domain when the RBD is in the up conformation (i.e., the active state). The increased mobility of the loop harboring the multibasic proteolytic site appears to increase the accessibility of the furin-like domain to the solvent and fixes the position of R685. This likely facilitates the site recognition by the protease and promotes the subsequent cleavage, leading to an increased rate of spike protein priming. The furin-like cleavage site is an evolutionary novelty in SARS-CoV-2 as compared with its close CoV relatives (Andersen et al. 2020; Xiao et al. 2020) and has been reported to play a crucial role for efficient cell–cell transmission (Hoffmann, Kleine-Weber, and Pöhlmann 2020; Hoffmann, Kleine-Weber, Schroeder, et al. 2020; Ou et al. 2020). In laboratory experiments, deletion of the S1/S2 motif resulted in a spike protein that was no longer able to induce syncytium formation whereas modification of the S1/S2 motif with a more efficient one (alanine-to-lysine substitution: RRAR -> RRKR) strongly increased syncytium formation potentially enhancing pathogenicity (Hoffmann, Kleine-Weber, and Pöhlmann 2020). Based on these data, our results argue for a refined efficiency of infection mechanism in the G614 variant.
The G614 Has a Long-Range Effect on the RBD Dynamics
A more in-depth inspection of the essential dynamics of the two protein variants detects a long-range effect of the D614G substitution, affecting the sampling of the RBD and its dynamical connection with other functional domains. To filter out the noise given by minor movements that could hamper the analyses of the main motions that dictate the large conformational movement of a protein/domain (D’Annessa et al. 2018, 2019), we decomposed the whole protein motion in its principal components (PCA) and discuss here the most relevant motions described by the first three eigenvectors. The two variants of the spike display clear differences in the amplitude of the conformational space sampled by the RBD in the up conformation along the first three eigenvectors (fig. 4A and supplementary fig. S12, Supplementary Material online), together describing more than 50% of the total motion. Conversely, the essential dynamics is largely similar between the two variants for the chains with the RBD in down conformation (data not shown). In general, the motion of the whole S1 subdomain, including the RBD, is more spread out in D614, whereas the same region in G614 has a more confined motion. However, the receptor-binding motif (RBM residues 435–506)—the apical region of the RBD directly deputed to bind the ACE2 receptor—shows a larger movement. This movement of the RBM would also allow it to move far apart from the N-terminal domain (NTD), as evidenced by the increase in distance between these two domains in G614 (fig. 4B and supplementary movie M1, Supplementary Material online). Conversely, even if the NTD and RBD in D614 appear to sample a larger space, their relative position does not vary during the course of the simulation (fig. 4B and supplementary movie M1, Supplementary Material online).
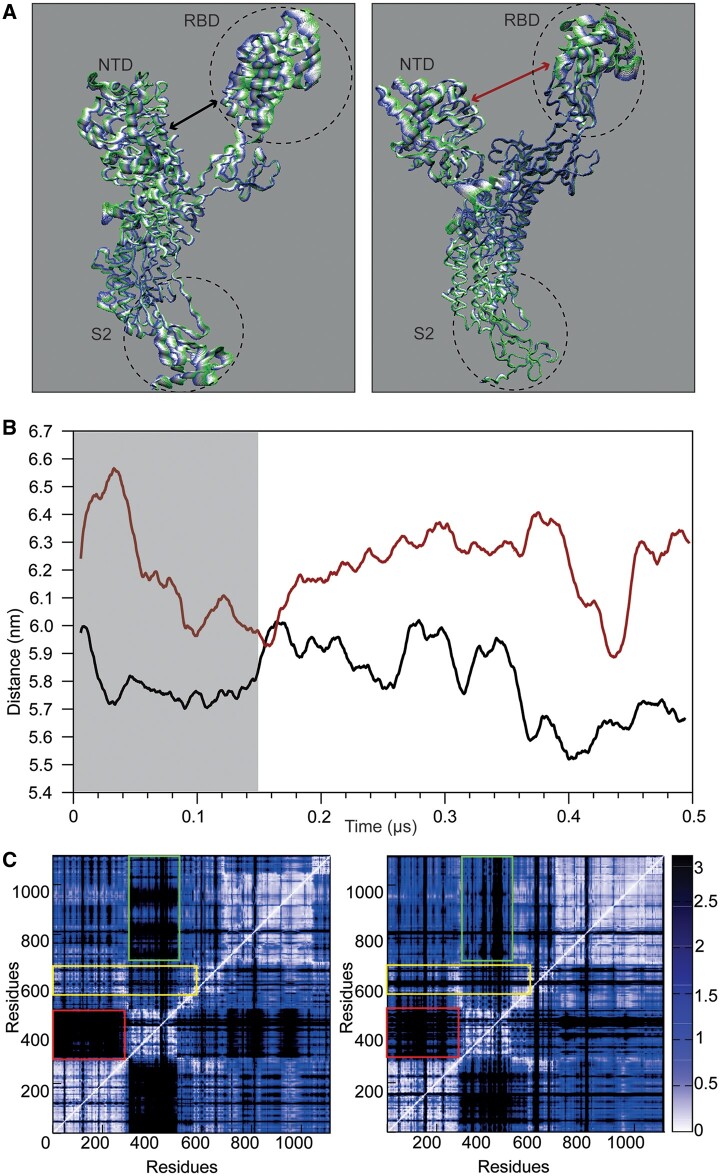
Fig. 4.
Principal component analysis of chain. (A) Projection of the motion along the first eigenvector for the chain A of D614 (left) and G614 (right). The color code from green to blue and the thickness of the tube identify the amplitude of the motion. (B) Evolution of the distance calculated between the center of mass of the NTD and the RBD in D614 (black) and G614 (red) as a function of time. (C) Distance fluctuation matrices calculated for chain A in D614 (left) and G614 (right). The rectangles underline the regions with differences between the two proteins.
The differences in the interdomain coordination can be better highlighted by plotting the matrix reporting the distance fluctuation (DF) between pairs or residues (fig. 4C). In this way, we can filter out the regions that are connected dynamically and functionally by following the variation in the relative distance among selected residues (Morra et al. 2012; D’Annessa et al. 2019). Basically, a variation in the pairs distance is an index of low dynamical connection between the two residues in the pair. In D614 the dynamical connection between the NTD and the RBD is low (fig. 4C, left panel, red rectangle), as well as that between the RBD and the S2 domain (fig. 4C, left panel, green rectangle). On the contrary, the NTD–RBD connection becomes stronger in the G614 variant, once again supporting the conclusion that the two domains are dynamically and functionally more connected (fig. 4C, right panel, red rectangle). In this case, a very strong connection also appears between the RBD and the S2 domain (fig. 4C, right panel, green rectangle), highlighting a stronger functional interdomain connection in the G614 variant. Interestingly, in G614 residues 675–692 are poorly connected with the rest of the protein, when compared with D614 (fig. 4C, yellow rectangles), once again consistent with a peculiar motion of the furin-like domain in the G614 variant.
We conclude that the dynamics of the RBD domain is different in the two 614 variants, with the domain in G614 exploring a more open conformation, suggesting a long-range effect of the D to G substitution that likely increases the efficient interaction of RBD with the ACE2 receptor. This is in line with the recent cryo-EM structure of the cognate SARS-Cov spike protein bound to ACE2 provided by Song et al. (2018) where the ACE2-bound RBD is shown as more open when compared with the ACE2-free RBD in the up conformation. This increased opening of the RBD domain with respect to the vertical axis of the trimer is a prerequisite for allowing the proper interaction with the receptor and for further transition from pre- to postfusion conformation (Song et al. 2018). Importantly, our results are in full agreement with the recent structural and functional analysis of the G614 Sars-Cov-2 spike protein by cryo-EM, showing that this mutation directly affects the dynamics of the RBD (Yurkovetskiy et al. 2020). In particular, the RBD shows extremely increased flexibility, precluding a high resolution in the cryo-EM map, and allowing the Spike protein to achieve a more open conformation via an increase in the S1-NTD and S1-INT distances, consistent with our analyses (fig. 4A and B). Such facilitation of the conformational switch, likely associated with a lower energetic barrier, could be the cause of a 4–13 times higher infectivity, depending on the type of cell targeted, of the G614 as compared with the D614 variant (Plante et al. 2020; Yurkovetskiy et al. 2020). Higher infectivity is unlikely to be due to an increased affinity of the protein for the human receptor as Yurkovetskiy et al. (2020) also detect a 5.7 times lower binding affinity of the G614 variant for ACE2.
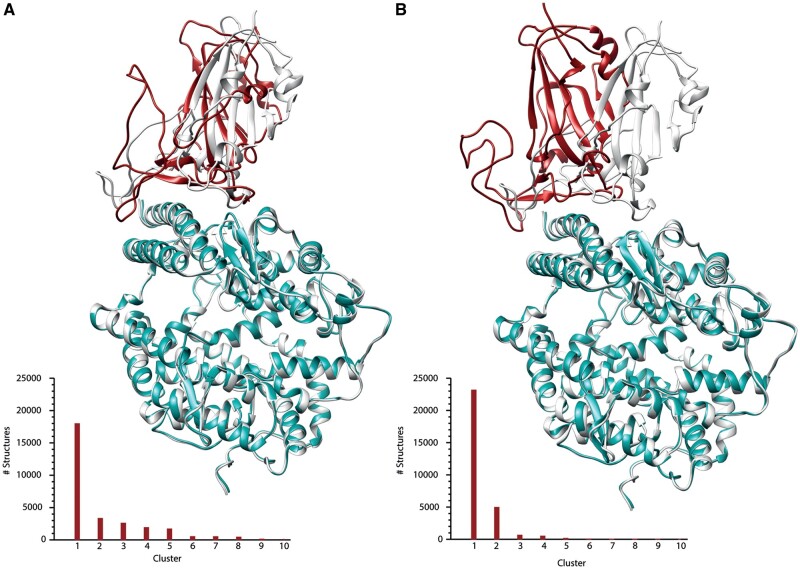
We confirmed the lower affinity by providing an atomistic model of the RBD–ACE2 interaction interface for the two variants, showing a slightly lower binding affinity of the G614_RBD for the ACE2 receptor, compared with the D614_RBD. Comparing the structures of the best docking complex obtained for the two variants with the X-ray structure of the RBD–ACE2 complex of the D614 ancestral variant (Lan et al. 2020), we show that the G614 variant is unable to fully reproduce the RBD–ACE2 binding mode captured by X-ray crystallography, whereas D614_RBD contacts ACE2 with the same structural features observed in the experimental structure (fig. 5A and B, respectively). Indeed, the D614_RBD–ACE2 and G614_RBD–ACE2 display a root mean square deviation (RMSD) value calculated for the Cα atoms of 1.8 and 3.2 Å with the X-ray structure, respectively. Furthermore, the Haddock score, which resembles the binding affinity (see Materials and Methods section for definition of the Haddock score), has a value of −131 for the D614 variant as compared with −105 for the G614 variant, suggesting a lower affinity of the latter for the ACE2 receptor, in line with what has been recently reported (Yurkovetskiy et al. 2020). Our results support the hypothesis that the basis of the advantage of the G614 variant may be the establishment of a faster but not stronger interaction with the receptor, which allows the virus to more easily engage the host membrane.
Fig. 5.
Cluster analysis and molecular docking of RBD–ACE2 interaction. Superposition of the RBD–ACE2 X-ray structure (Lan et al. 2020) with the best complex obtained through docking for the D614_RBD–ACE2 (A) and G614_RBD–ACE2 (B). The X-ray structure is shown in gray; the RBD and the ACE2 from the docking calculations are shown in red and cyan, respectively. Structures belonging to each cluster detected in the last 350 ns for the both variants (histograms at the bottom-left of each panel) show that the structural ensemble of both RBD variants is mainly represented by a highly populated family of structures.
Conclusions
The G614 spike variant rose to fixation in the genomic samples from the vast majority of countries in the world (fig. 2B). By statistically controlling for random genetic drift within geographical areas and for migration among different areas, we showed that the available data are compatible with a selective advantage of G614. Moreover, the increasing frequencies in the genomic samples are mirrored by similar trends in the actual virus populations reconstructed by a coalescent-based approach, which argues against an overrepresentation of G614 in the genomic samples, putatively due to enhanced severity of the COVID-19 symptoms, as also directly assessed in patients from UK hospitals (Volz et al. 2020). Even if our results are robust to different metrics designed to control for gene flow among geographic areas, we caution that complex epidemiological patterns may be not fully accounted for by our relatively simplistic models of SARS-CoV-2 population dynamics. These models are designed to capture the current knowledge about the unfolding, yet not fully understood, virus global outbreak. However, as the understanding of the parameters governing this epidemic improves, more direct approaches (i.e., simulations) can be deployed to test for a selective advantage of any given variant.
The structural analysis of the spike protein revealed the crucial role of position 614 in interacting with both the S1/S2 furin-like and the RBD. The substitution in this position of an aspartic acid (D), carrying a negatively charged side chain, with a glycine (G) strongly affects the dynamics of the spike functional domains at both short and long ranges, allowing it to sample novel structural conformations, and, we suggest, being the underlying cause of the frequency increase of this SARS-Cov-2 variant. In particular, the long-range effect of the D614G substitution influences the internal dynamics of the activated RBD and its relative orientation, allowing it to acquire a more open conformation. The increased opening of the RBD conformation appears not to increase affinity toward ACE2 receptor, but instead may reflect an increased suitability of the G614 variant RBD to more easily switch from an inactive (i.e., down) to an active (i.e., up) conformation, consistent with our docking analysis and as suggested recently (Yurkovetskiy et al. 2020). The presence of a glycine in position 614 may serve to lower the energetic barrier associated with the down-to-up conformational change of the RBD. Nevertheless, the stronger dynamical connection of the RBD with the S2 domain could favor removal of the steric restraints on helix linker 2, which would better trigger the release of the S1 subunits, allowing the extension of prefusion S2 helixes to form the postfusion S2 long helix bundle (Song et al. 2018).
Finally, compared with the ancestral form, the G614 variant shows a marked conformational plasticity of the furin-like domain (i.e., short-range effect), also increasing the volume of the cavity surrounding the cleavage site. These structural and dynamical features may possibly favor recognition of the furin-like domain by the protease and as a consequence improve the efficiency of the proteolytic cleavage that represents a crucial step for host cell infection. However, what appears to be the evolutionary strength of the virus in terms of invasiveness could be transformed into its weakness. Indeed, the peculiar properties of the furin-like domain in the G614 variant, such as the widening of the cavity, could be exploited for the rational design of drug molecules that are able to bind it and compete for the interaction of the spike with the protease, in essence specifically blocking the invasiveness of this SARS-CoV-2 variant. In addition, as the G614 could not be the only cause of the selective advantage of this SARS-CoV-2 variant, the structural and functional changes caused by the other nonsynonymous substitution in the same linkage group (i.e., the C14408T in the nsp12 protein) should be thoroughly investigated to shed light on any potentially synergistic effect on the virus spread.
Materials and Methods
Models of Temporal Variation of G614 in the Genomic Samples
Complete high-coverage whole-genome sequences of SARS-CoV-2 from European, US, Australian, and Chinese isolates were downloaded from the GISAID EpiFluTM Database on September 27, 2020. A total of 71,124 sequences were aligned to the Wuhan-Hu-1 SARS-CoV-2 reference sequence (MN908947.3) (Wu et al. 2020) using mafft (Katoh and Toh 2008). Alignment was parsed using a custom python script to discard sequences with more than 500 missing bases and to extract polymorphic sites.
We modeled the probability that a sampled viral genomic sequence has a G at spike amino acid position 614 as a function of time by fitting GLMM and phylogenetic generalized linear mixed models (pGLMM) with binomial error structure, including random intercepts and slopes for geographic areas (countries, US states, or Chinese provinces), and covariance matrices based on the genetic or spatial similarity between viral samples from each pair of areas. The rationale for this model structure is that geographic areas can be seen as local, but highly interconnected, genetic drift units (i.e., demes) of the global outbreak where genetic alleles are randomly, yet nonindependently, sorted within each area.
Specifically, we modeled the covariance structures for nonindependence among geographic regions as follows: 1) GLMMs using the covariance in sample allele frequencies as in Pickrell and Pritchard (2012), where the correction for branch length was adapted to haploid genomes; as this model design does not require a phylogenetic tree, it corresponds to an island model where gene flow is continuous between populations; 2) phylogenetic GLMMs using covariance matrices obtained by FST-based phylogenetic trees with the VCV function of the R package ape (Goudet 2005) (FST values were calculated using the function pairwise.fst, R package hierfstat v0.04-22); and 3) spatial models using the coordinates of population centroids of each geographic area (Hall et al. 2019). In all of these models, random slopes of day within geographic areas capture random variation of allele frequency change among demes (i.e., random drift within each deme and, in principle, possible differences in the strength of selection), covariance matrices capture the relative amount of drift that is shared between demes (due to migration, i.e., exchange of viral lineages), and the fixed effect of sampling day represents the change in frequency that is not explained by genetic drift within areas or is by connectivity among areas. Genetic covariance matrices were computed on the full set of sequence data or on subsets including only sequences with the G614 allele. Moreover, all models were tested both on all sequences and on a subset of sequences collected in April 2020, where gene flow among countries was restricted due to travel ban among most of the countries in the world. Models including genetic covariance matrices were fitted in a Bayesian framework using the R package brms (Bürkner 2017), with four chains of 4,000 iterations. Spatial models were estimated with the R package spaMM v3.2.0 (Rousset and Ferdy 2014) fitting a Matern covariance matrix using the latitude and longitude of the geographic areas. Statistical significance of the logistic slope for all models described above was assessed by calculating 95% confidence intervals. For models fitted in brms, confidence limits were computed as the 2.5% and 97.5% quantiles from the posterior sample. For models fitted in spaMM, the confint function was used, which applies an approximation of the profile likelihood. Covariance matrices used for the different models are represented graphically in supplementary figures S1–S6, Supplementary Material online. As remarked by Volz et al. (2020), under the assumptions of exponential growth for the whole population and selection being the sole driver of allele frequency change, a logistic slope for the relative frequency of one allele over time translates to a selection coefficient (s). Following Volz et al. (2020), we computed selection coefficients for the global population and for each geographic area as s = ρ/r, where ρ is the logistic slope and r the exponential growth rate for the viral population. We considered two extreme values for r (rmin, rmax), calculated by numerical approximation assuming a serial interval of 6.5 days and basic reproduction numbers 2.0 and 3.5 (Flaxman et al. 2020).
We also explored the possibility that a single founder effect event could have led to a dramatic increase in G614 in a single area that would have later contributed genomes worldwide with a disproportionate number of migrants. Specifically, we hypothesized that countries which saw an early emergence of G614 might be the most representative source populations contributing viral genomes globally. Iceland and Belgium are the countries with the highest estimated frequency of G614 on February 1, 2020, and Belgium had one of the largest per capita prevalence of COVID-19, whereas Italy reported a large number of cases early in the pandemic. We built distinct source–sink linear models for each of these three countries, assuming that, after February 1, 2020, the country of interest acted as source and all the other geographic areas in the world as sinks. In our source–sink models, the logistic slope kj of G614 over time in the sink area j is assumed to be proportional to mij(pi − pj(0)), where mij is the migration rate from the source area i to sink area j, pj(0) the frequency of G614 in sink area j at time 0, and pi the frequency of G614 in the source i. To see this, notice that, for a sink population j with allele frequency pj(t) receiving a proportion of migrants mij from a source population i with frequency pi, the allele frequency at time t + 1 is pj(t + 1) = (1 − mij)·pj(t) + mijpi. Thus, pj (t) = (1 − mij)t·(pj(0) − pi) + pi. Through first-order Taylor approximations of pj(t) (assuming, realistically, that migration rate per day mij is small) and of a logistic equation with slope k around its midpoint, we obtain that k ∼ mij·(y − pj(0)). To estimate mj·(pi − pj(0)), we used pi and pj(0) as predicted by our main model (pGLMM on full data set and shared drift modeled by covariance matrix, see above) on February 1, 2020 (before G614 increased globally in frequency) for Iceland and Belgium. For Italy, which had a high number of cases but a lower predicted frequency of G614 on February 1, 2020, we assumed a frequency of 100%. We used (1 − FSTij)/FSTij as a proxy of mij. Note, however, that (1 − FSTij)/FSTij ∼ Njmij, rather than mij itself. Thus, variations in Nj and stochastic factors affecting the number of migrants across geographic areas were modeled as a Gaussian random effect on the slope for each state. We also included, in the source–sink linear models, random intercepts for each geographic area, representing the variation of the time of contact between the source and the different sink areas. The structure of the source–sink models was thus p ∼ 1 + Z(xij·day) + (1 + Z(xij·day) | geographic_area), where p is the relative frequency of the G614 allele, Z indicates that a predictor was Z transformed, and xij was set as xij = (pi − pj(0))·(1 − FSTij)/FSTij, with pIceland = 0.932, pBelgium = 0.751, and pItaly = 1. Source–sink models were compared with a model in which only the linear predictor day is included (i.e., where xij = 1) by means of AIC and BIC, to assess whether single founder effect followed by widespread dissemination of the G614 variant may explain its steady worldwide increase better than time alone (a significant improvement of the model performance when xij ≠ 1 would imply that source–sink mechanisms contribute in explaining the increase in frequency of the G614 variant).
Coalescent-Based Inference of A and G614 Variants Diffusion in the Population
Whole-genome viral sequences were grouped according to the nucleotidic allele at position 23403 (whether A or G, corresponding to the D614 and G614 variants, respectively) and then by geographic area, resulting in two alignments per area. Selected pairs of alignments with at least 30 sequences for each of the two alleles were retained for downstream analyses (Australia, Belgium, England, Iceland, Netherlands, Spain, the United States—NY, the United States—WA, the United States—WI, and Wales). The maximum number of sequences to be used in the coalescent-based analysis was limited to 250, which were randomly selected when needed. Three independent replicates of randomly selected sequences were run and checked for consistency of the results.
The demographic history of the G614 monophyletic clade and the overall remaining SARS-CoV-2 phylogenetic tree (D614) was reconstructed in each geographic area using the Bayesian Skyline plot analyses as implemented in Beast v2.6 (Bouckaert et al. 2019). For each alignment, we prepared the input file by setting the tips dates as days before the most recent sequence (available as “Collection date” in the GISAID metadata), an HKY substitution model, a strict clock model, a coalescent BSP as tree prior, and 100,000,000 iterations for the Markov chain Monte Carlo. We ran three replicates for each alignment and checked their convergence in Tracer 1.8 (Rambaut et al. 2018). Bayesian Skyline plot analyses (Drummond et al. 2005) were run in Tracer and results exported as tabular values. We then calculated the relative frequency through time of the G614 variant in the population by dividing the median values of the estimated effective population size of the G614 by the sum of the median values of the estimated effective population sizes of A and G614 (Ne(A) and Ne(G), respectively). The confidence interval of the estimated relative frequency of the G614 was calculated using the 95% confidence intervals for the individual A and G614 BSP: for the upper boundary = 97.5% Ne(G)/(97.5% Ne(G) + 2.5% Ne(A)); lower boundary = 2.5% Ne(G)/(2.5% Ne(G) + 97.5% Ne(A)). Plots were drawn in R and python using standard plotting functions and libraries. Note that this procedure is conservative and might result in a substantial overestimate of the uncertainty.
MD Simulations of D614 and G614 Variants
The starting structure of the Spike trimeric complex with one RBD in up and two in down conformation was taken from the structure deposited in the protein data bank with code 6VSB (Wrapp et al. 2020). Missing residues, mainly belonging to loops regions, were reconstructed using the SwissModel web server (https://swissmodel.expasy.org/, last accessed on 15/10/2020). Because of the lack of a reliable template to model the 3D arrangement of the HR2 segment with respect to the rest of the protein, and thus to model the transmembrane region, the model covers residue 27-1146. All the N-acetylglucosamine residues present in the cryo EM structure bound to asparagines residues were retained in the system. The D to G mutation in position 614 was introduced with the Chimera program (Pettersen et al. 2004) and the structure obtained was further minimized. Topologies of the two systems were built using tleap with the AMBER14 force field (Case et al. 2014). Each protein was then placed in a triclinic simulative box filled with TIP3P water molecules (Jorgensen et al. 1983). Addition of sodium counterions rendered the systems electroneutral; each system consisted of approximately 555,000 atoms.
The simulations were carried out with amber14 using pmemd. CUDA (Case et al. 2014). The systems were first minimized with 10,000 steps of steepest descent followed by 10,000 steps of conjugate gradient. Relaxation of water molecules and thermalization of the system in NPT environment were run for 1.2 ns at 1-fs time step. In detail, six runs of 200 ps each were carried out by increasing the temperature of 50 K at each step, starting from 50 to 300 K. The systems were then simulated with a 2-fs time step for 500 ns each in periodic boundary conditions, using a cut-off of 8 Å for the evaluation of short-range nonbonded interactions and the Particle Mesh Ewald method (Cheatham et al. 1995) for the long-range electrostatic interactions. The temperature was kept constant at 300 K with Langevin dynamics (Ceriotti et al. 2009) and pressure fixed at 1 Atmosphere through the Langevin piston method (Feller et al. 1995). The bond lengths of solute and water molecules were restrained with the SHAKE (Ryckaert et al. 1977) and SETTLE algorithms (Miyamoto and Kollman 1992), respectively. As stated before, the transmembrane region is lacking. In order to mimic the binding of the spike trimer on the viral membrane, we applied a force of 1,000 kj on the last four residues (1143–1146) to anchor the protein. Analyses were carried out using Gromacs 5 package (Hess et al. 2008) or with VMD (Humphrey et al. 1996) and custom code.
As reported in supplementary figure S11C and D, Supplementary Material online, both proteins largely deviate from their starting conformation, reaching an RMSD value of approximately 0.4 nm. This was, however, expected and due to the fact that the starting configuration used to carry out the simulations comes from cryo-EM and was solved at a resolution of 3.46 Å. It is then reasonable that once hydrated, the proteins undergo sudden conformational changes to relax the structure. However, the RMSD in both cases reaches a plateau at around 0.15 µs, meaning that the two proteins find the stability, and for this reason all analyses reported here have been performed on the last 0.35 µs of simulative time.
Volumes were computed using POVME (Wagner et al. 2017). An inclusion sphere with a radius of 11 Å was manually placed between the furinic loop and the near S2 domain after careful optimization in VMD (Humphrey et al. 1996).
The RMSDs from the starting structure has been calculated by:
where M is the sum of atomic masses, is the mass of atom i, and t = 0 refers to the selected reference structure. The instantaneous configurations at time t are obtained by removing the global translations and rotations.
The per-residue root mean square fluctuations (RMSF) have been computed by using the following definition:
where the averages have been calculated over the equilibrated MD trajectories.
Principal component analysis for the bound and unbound trajectories was carried out on the 3N × 3N Cartesian displacement matrix whose elements are calculated as:
N being the number of Cα atoms and qi the (mass-weighted) displacement of the ith Cα atoms from the reference value (after removal of rotational and translational degrees of freedom).
The matrix of the DF was computed as:
being dij the (time-dependent) distance of the Cα atoms between amino acids i and j and 〈 〉 the time average over the trajectory. DF is independent upon translations and rotations of the molecules and thus on the choice of a protein reference structure.
Docking Calculations for the RBD–ACE2 Interaction Prediction for the D614 and G614 Variants
The structures of the RBD in the two simulations (i.e., D614 and G614) have been clustered in order to extract for each RBD variant the most representative conformation to be used for docking calculations. Clustering based on the RMSD among the sampled structures has been carried out with Gromacs 5 package (Hess et al. 2008) using the GROMOS clustering method. As shown in figure 5, in the last 350 ns of sampling the structural ensemble of both RBD variants is mainly represented by a highly populated family of structures. The representative structure of the most-populated family has been selected for further calculations, whereas the structure of the ACE2 receptor has been extracted from the RBD–ACE2 crystal structure deposited in the protein data bank with code 6M0J (Lan et al. 2020). The docking has been carried out using the web interface of the HADDOCK software, particularly well performing in protein–protein docking predictions (de Vries et al. 2010; Di Marino et al. 2015).
The HADDOCK pipeline is built on a knowledge-based algorithm in which the docking is driven by experimental evidence; the residues at the RBD–ACE2 interface in the X-ray structure were used as active residues (AIRs) to drive the docking calculations. For each docking, the poses of the complexes obtained are classified based on the HADDOCK score, resulting from the sum of different energetic contributions to the binding, for example, hydrogen bonds, van der Waals and electrostatic interaction, buried solvent accessible surface. In principle, the lowest is the HADDOCK score, the highest is the predicted affinity between ligand and receptor.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
All SARS-CoV-2 genome sequences used in this study were downloaded from the GISAID EpiFlu Database, www.gisaid.org (accessed on September 27, 2020). We thank Professor Giorgio Colombo for providing the code for the calculation of the distance fluctuation matrices. The work was partially supported by funding to D.D.M. by Fondazione Marche.
Author Contributions
E.T. conceived the idea with D.D.M.; E.T., P.G., Fa.M., and G.B. designed the analyses to test for the selective advantage of 614G variant; P.G. and Fa.M. implemented the population dynamics analyses; E.T. and P.G. implemented the population demographic reconstructions; I.D. run the molecular dynamics simulations and carried out the analyses of the trajectories together with S.M.; E.T., P.G., Fa.M., F.C., Fi.M. G.B., I.D., and D.D.M. discussed the results and wrote the manuscript.
Data Availability
Genomic data alignment and genomic variants tested in this study are available here: https://doi.org/10.6084/m9.figshare.13493178.v1 and https://doi.org/10.6084/m9.figshare.13498428.v1. Code for GLMM and pGLMM analyses presented in this study is available here: https://github.com/fabrimafe/G614_SARS-Cov2.
References
- Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF.. 2020. The proximal origin of SARS-CoV-2. Nat Med. 26(4):450–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra-Flores M, Cardozo T.. 2020. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int J Clin Pract. 74(8):e13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S, Millet JK, Licitra BN, Whittaker GR.. 2012. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4(6):1011–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JD, Jones S, Drebot MA, Andonov A, Sabara M, Yuan XY, Weingartl H, Fernando L, Marszal P, Gren J, et al. 2004. Development and characterisation of neutralising monoclonal antibody to the SARS-coronavirus. J Virol Methods. 120(1):87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N, et al. 2019. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 15:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brufsky A. 2020. Distinct viral clades of SARS-CoV-2: implications for modeling of viral spread. J Med Virol. 92(9):1386–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürkner PC. 2017. brms: an R package for Bayesian multilevel models using Stan. J Stat Softw. 80(1):1–28 [Google Scholar]
- Case DA, Berryman JT, Betz RM, Cai Q, Cerutti DS, Cheatham TE III, Darden TA, Duke RE, Gohlke H, Goetz AW, et al. 2014. The Amber Molecular Dynamics Package.Available from: http://ambermd.org/ [Google Scholar]
- Ceriotti M, Bussi G, Parrinello M.. 2009. Langevin equation with colored noise for constant-temperature molecular dynamics simulations. Phys Rev Lett. 102(2):020601. [DOI] [PubMed] [Google Scholar]
- Cheatham TE, Miller JL, Fox T, Darden TA, Kollman PA.. 1995. Molecular dynamics simulations on solvated biomolecular systems: the particle mesh Ewald method leads to stable trajectories of DNA, RNA, and proteins. J Am Chem Soc. 117(14):4193–4194. [Google Scholar]
- Chiara M, Horner DS, Pesole G.. 2020. Comparative genomics suggests limited variability and similar evolutionary patterns between major clades of SARS-Cov-2. bioRxiv. 10.1101/2020.03.30.016790. [DOI]
- Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E.. 2020. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 176:104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Annessa I, Gandaglia A, Brivio E, Stefanelli G, Frasca A, Landsberger N, Di Marino D.. 2018. Tyr120Asp mutation alters domain flexibility and dynamics of MeCP2 DNA binding domain leading to impaired DNA interaction: atomistic characterization of a Rett syndrome causing mutation. Biochim Biophys Acta Gen Subj. 1862(5):1180–1189 [DOI] [PubMed] [Google Scholar]
- D’Annessa I, Raniolo S, Limongelli V, Di Marino D, Colombo G.. 2019. Ligand binding, unbinding, and allosteric effects: deciphering small-molecule modulation of HSP90. J Chem Theory Comput. 15(11):6368–6381. [DOI] [PubMed] [Google Scholar]
- D’Arienzo M, Coniglio A.. 2020. Assessment of the SARS-CoV-2 basic reproduction number, R0, based on the early phase of COVID-19 outbreak in Italy. Biosaf Health. 2(2):57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries SJ, van Dijk M, Bonvin AMJJ.. 2010. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 5(5):883–897. [DOI] [PubMed] [Google Scholar]
- Di Marino D, Chillemi G, De Rubeis S, Tramontano A, Achsel T, Bagni C.. 2015. MD and docking studies reveal that the functional switch of CYFIP1 is mediated by a butterfly-like motion. J Chem Theory Comput. 11(7):3401–3410. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG.. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 22(5):1185–1192. [DOI] [PubMed] [Google Scholar]
- Feller SE, Zhang Y, Pastor RW, Brooks BR.. 1995. Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys. 103(11):4613–4621. [Google Scholar]
- Flaxman S, Mishra S, Gandy A, Unwin HJT, Mellan TA, Coupland H, Whittaker C, Zhu H, Berah T, Eaton JW, et al. ; Imperial College COVID-19 Response Team. 2020. Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature 584(7820):257–261. [DOI] [PubMed] [Google Scholar]
- Goudet J. 2005. HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes. 5(1):184–186 [Google Scholar]
- Hall O, Bustos MFA, Olén NB, Niedomysl T.. 2019. Population centroids of the world administrative units from nighttime lights 1992-2013. Sci Data. 6(1):1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JF, Peng GW, Min J, Yu DW, Liang WJ, Zhang SY, Xu RH, Zheng HY, Wu XW, Xu J, et al. 2004. Molecular evolution of the SARS coronavirus, during the course of the SARS epidemic in China. Science (80-.). 303:1666–1669. [DOI] [PubMed] [Google Scholar]
- Hess B, Kutzner C, Van Der Spoel D, Lindahl E.. 2008. GRGMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 4(3):435–447. [DOI] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Pöhlmann S.. 2020. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 78(4):779–784.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, et al. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181(2):271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K.. 1996. VMD: visual molecular dynamics. J Mol Graph. 14(1):33–8 [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML.. 1983. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 79(2):926–935. [Google Scholar]
- Katoh K, Toh H.. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 9(4):286–298. [DOI] [PubMed] [Google Scholar]
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, et al. 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182(4):812–827.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, et al. 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581(7807):215–220. [DOI] [PubMed] [Google Scholar]
- Li F. 2016. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 3(1):237–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Xiao G, Chen Y, He Y, Niu J, Escalante CR, Xiong H, Farmar J, Debnath AK, Tien P, et al. 2004. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 363(9413):938–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S, Kollman PA.. 1992. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem. 13(8):952–962. [Google Scholar]
- Morra G, Potestio R, Micheletti C, Colombo G.. 2012. Corresponding functional dynamics across the Hsp90 chaperone family: insights from a multiscale analysis of MD simulations. PLoS Comput Biol. 8:e1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, et al. 2020. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 11:1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachetti M, Marini B, Benedetti F, Giudici F, Mauro E, Storici P, Masciovecchio C, Angeletti S, Ciccozzi M, Gallo RC, et al. 2020. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 18:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak JE, Sharon C, Satkunarajah M, Auperin TC, Cameron CM, Kelvin DJ, Seetharaman J, Cochrane A, Plummer FA, Berry JD, et al. 2009. Structural insights into immune recognition of the severe acute respiratory syndrome coronavirus S protein receptor binding domain. J Mol Biol. 388(4):815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC, Kugathasan R, Penn R, Brown JC, Sanchez-David RY, et al. 2020. The furin cleavage site of SARS-CoV-2 spike protein is a key determinant for transmission due to enhanced replication in airway cells. bioRxiv. 10.1101/2020.09.30.318311 [DOI] [PubMed]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE.. 2004. UCSF Chimera - a visualization system for exploratory research and analysis. J Comput Chem. 25(13):1605–1612. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK.. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8(11):e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, Zhang X, Muruato AE, Zou J, Fontes-Garfias CR, et al. 2020. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 10.1038/s41586-020-2895-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA.. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 67(5):901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F, Ferdy JB.. 2014. Testing environmental and genetic effects in the presence of spatial autocorrelation. Ecography 37(8):781–790. [Google Scholar]
- Ryckaert JP, Ciccotti G, Berendsen HJC.. 1977. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 23(3):327–341. [Google Scholar]
- Simmons G, Zmora P, Gierer S, Heurich A, Pöhlmann S.. 2013. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 100(3):605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Gui M, Wang X, Xiang Y.. 2018. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 14(8):e1007236-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilarou M, Alachiotis N, Garefalaki J, Beloukas A, Pavlidis P.. 2020. Population genomics insights into the recent evolution of SARS-CoV-2. bioRxiv. 10.1101/2020.04.21.054122 [DOI] [PMC free article] [PubMed]
- Volz E, Hill V, McCrone JT, Price A, Jorgensen D, O’Toole Á, Southgate J, Johnson R, Jackson B, Nascimento FF, et al. 2020. Evaluating the effects of SARS-CoV-2 Spike mutation D614G on transmissibility and pathogenicity. Cell. 184(1):64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JR, Sørensen J, Hensley N, Wong C, Zhu C, Perison T, Amaro RE.. 2017. POVME 3.0: software for mapping binding pocket flexibility. J Chem Theory Comput. 13(9):4584–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D.. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181(2):281–292.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Tortorici MA, Snijder J, Xiong X, Bosch BJ, Rey FA, Veesler D.. 2017. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci U S A. 114(42):11157–11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JM, Whittaker GR.. 2016. Fusion of enveloped viruses in endosomes. Traffic 17(6):593–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO). 2020. Coronavirus disease 2019 Situation Report 51, March 11, 2020. World Health Organization. [Internet] 2019:2633. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS.. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367(6483):1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, et al. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579(7798):265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou JJ, Li N, Guo Y, Li X, Shen X, et al. 2020. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 583(7815):286–289. [DOI] [PubMed] [Google Scholar]
- Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, Wang Y, Baum A, Diehl WE, Dauphin A, Carbone C, et al. 2020. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 183(3):739–751.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798):270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Liu G, Meng K, Yang L, Meng G., . 2020. Rapid spread of mutant alleles in worldwide COVID-19 strains revealed by genome-wide SNP analysis. Research Square. 10.21203/rs.3.rs-23205/v1 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic data alignment and genomic variants tested in this study are available here: https://doi.org/10.6084/m9.figshare.13493178.v1 and https://doi.org/10.6084/m9.figshare.13498428.v1. Code for GLMM and pGLMM analyses presented in this study is available here: https://github.com/fabrimafe/G614_SARS-Cov2.