Figure 7.
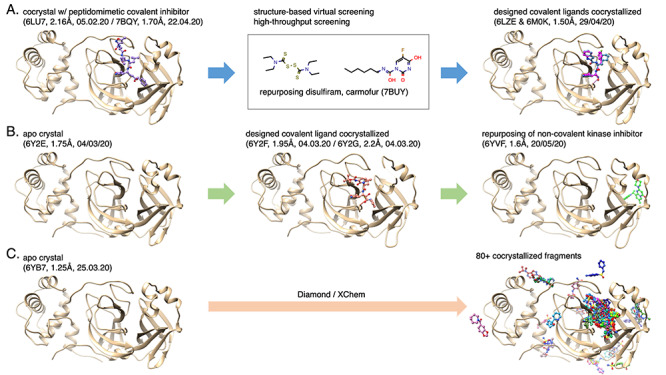
SARS-CoV-2 Main protease (3CLpro): earliest, advanced structure-based drug discovery routes (pdb entry, resolution, release date). The proteins are displayed in beige cartoon and the ligands in colored ball-and-sticks. (A) the first crystal of 3CLpro enabled the discovery of disulfiram and carmofur as potential drugs to repurpose through structure-based virtual and high-throughput screenings [10] as well as the design of peptidomimetic covalent ligands [159]; (B) an apo crystal was subject to structure-based design for covalent ligands [148] and recently brought rational for repurposing a non-covalent small molecule initially developed as a kinase inhibitor; (C) The XChem initiative started with the resolution of an apo structure allowing soaking experiments that led to more than 80 cocrystals with covalent and non-covalent fragments, most are located in the active site.
