Figure 5.
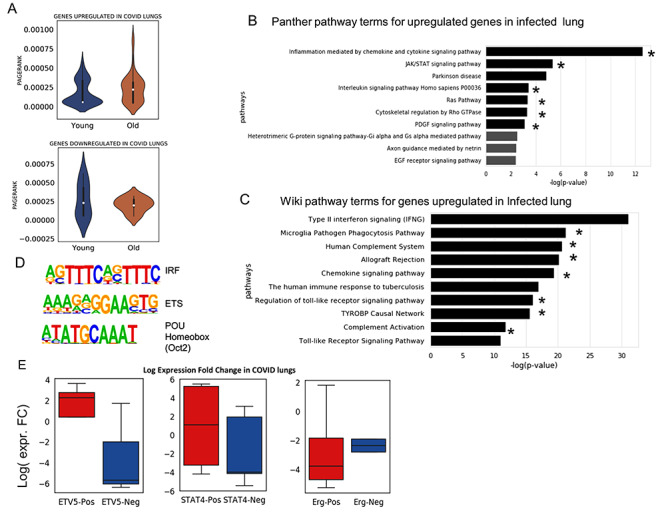
Analysis of gene expression profile in lungs infected with SARS-CoV-2 (COVID). (A) The distribution of PageRank of genes upregulated in COVID-infected lung false detection rate (FDR < 0.05) [37]. The PageRank is shown for network estimated using the scRNA-seq profile of young and old AT2 cells. On similar pattern PageRank is shown for genes downregulated FDR < 0.05 in COVID-infected lung. (B) Top 10 panther pathway enriched for genes upregulated in COVID-infected lung. The terms with * sign also have significant enrichment for genes with higher PageRank in old AT2 cells (C) Top 10 Wiki pathway terms enriched for genes upregulated in COVID-infected lung. The terms with * are also enriched p-value < 0.05 for genes with higher PageRank in old AT2 cells. (D) Top 3 motifs of known TF enriched in promoters of genes upregulated in COVID-infected lung. (E) Fold change of expression in the lung with COVID infection for genes positively and negatively correlated with TFs in old AT2 cells. The results are shown for 2 TFs Etv5 and Stat4, which has higher PageRank in old AT2 cells. As a control, the results are also shown for Erg, which have higher PageRank in young AT2 cells. Most of the genes, which had a positive correlation with Etv5 and Stat4 expression in old murine AT2 cells, were upregulated in COVID-infected lung, whereas for Erg the trend is the opposite. Genes positively correlated with ERG genes in old AT2 had more downregulation than genes with negative correlation. Such results hint that TFs whose influence (PageRank) increase during ageing could be involved activating or poising the genes upregulated in COVID infection.
