Figure 1. Syntaxin‐1 fragments rescue deficits in Munc18‐1 levels and solubility.
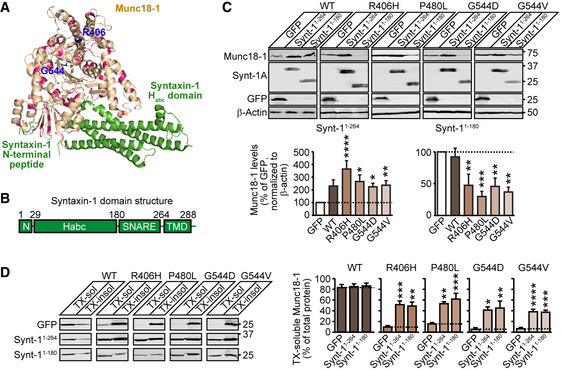
- Localization of disease‐causing missense mutations of Munc18‐1b in its tertiary structure (PDB code 4JEU) (Burkhardt et al, 2008), with annotation of residues and binding sites of syntaxin‐1 Habc domain and N terminus. Highlighted in blue are the residues 406 and 544 that we have analyzed in this study in detail.
- Syntaxin‐1 domain structure. The N terminus (residues 1‐28) binds to domain 1 of Munc18‐1 while the Habc domain (residues 28–180) and the SNARE domain (residues 180–264) bind in Munc18‐1’s central cleft. The transmembrane domain (TMD; residues 264–288) does not participate in Munc18‐1 binding.
- Total protein levels of Munc18‐1. HEK293T cells transfected with WT or mutant Munc18‐1 variants and either GFP, syntaxin‐11‐264 or syntaxin‐11‐180 were lysed and lysates were analyzed by quantitative immunoblotting to indicated proteins, normalized to β‐actin (Synt‐1A = syntaxin‐1A). Data are means ± SEM (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by Kruskal–Wallis test, followed by Dunn’s multiple comparison test; n = 11 independent experiments; exact P‐values are shown in Appendix Table S1).
- Solubility of Munc18‐1. HEK293T cells transfected as in (C) were solubilized in 0.1% Triton X‐100 (TX), and equal volumes of soluble and insoluble fractions were analyzed by quantitative immunoblotting. TX‐soluble Munc18‐1 was measured as percent of total Munc18‐1 by quantitative immunoblotting. Data are means ± SEM (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by Kruskal–Wallis test followed by Dunn’s multiple comparison test, or by one‐way ANOVA followed by Bonferroni post hoc test; n = 5–9 independent experiments; exact n and P values are shown in Appendix Table S1).
Source data are available online for this figure. Source data are available online for this figure.
