Figure 4. Bacteroides OM glycolipids signal through TLR4 to induce IFNβ.
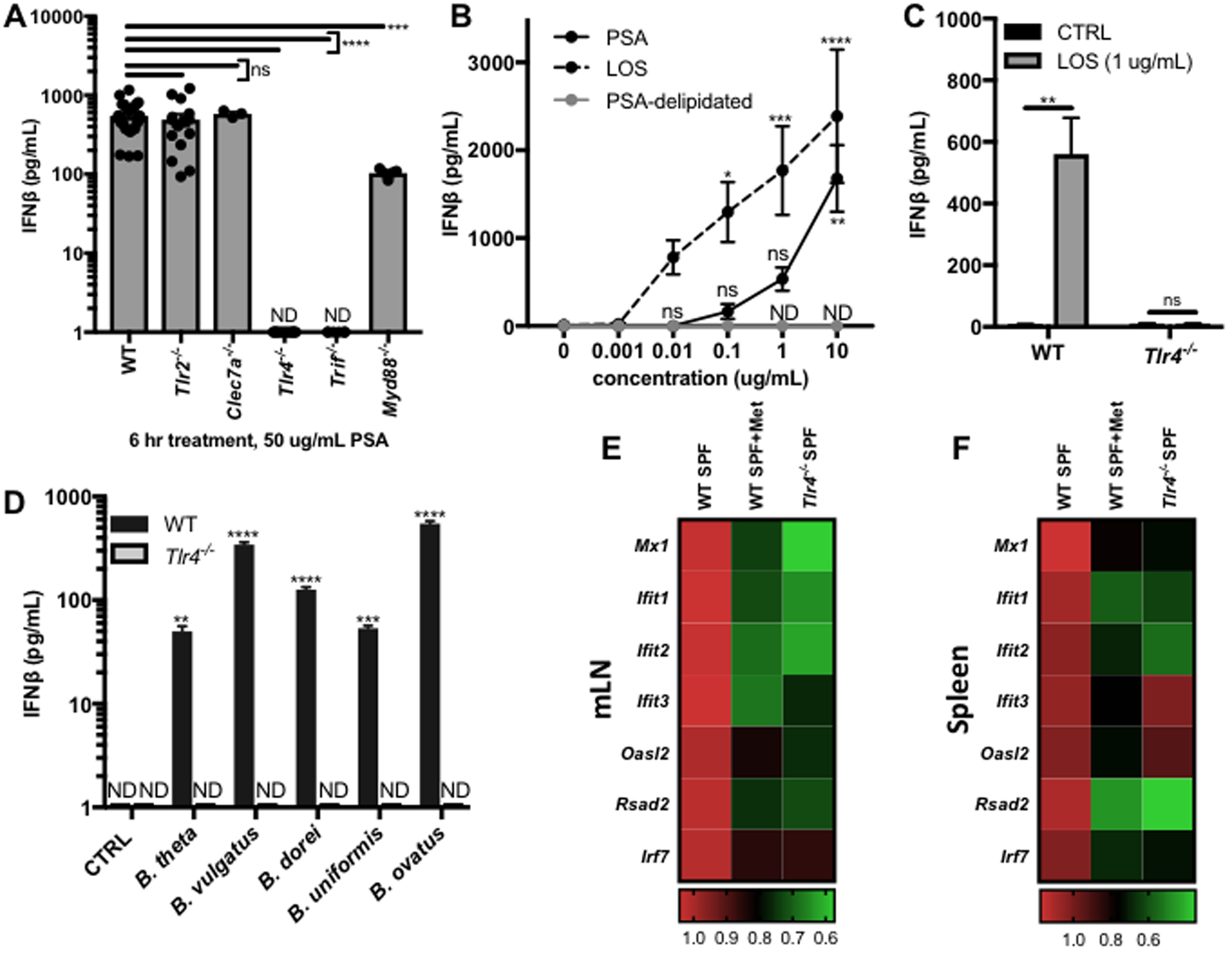
(A) BMDCs were differentiated from WT (N=27), Tlr2−/− (N=15), Clec7a−/− (N=3), Tlr4−/− (N=12), Trif−/− (N=6), or Myd88−/− (N=5) mice and treated with 50 ug/mL B. fragilis PSA for 6 hrs. IFNβ in the supernatants was measured by ELISA and normalized by subtracting vehicle control. (B) ELISA analysis of IFNβ in the supernatants of WT BMDCs treated for 6 hrs with a dose response of PSA (N=9), the LOS lipid domain of PSA (LOS, N=14), or delipidated PSA (PSA-delipidated, N=3, ND for all concentrations). ELISA analysis of IFNβ in the supernatants of WT or Tlr4−/− BMDCs treated for 6 hrs with (C) vehicle control (CTRL, N=8), 1 ug/mL LOS (N=8) or (D) 100 ug/mL Bacteroides OM extracts (N=3 per condition). (A-D) Data represents average +/− SEM. (E,F) RNA isolation and qRT-PCR analysis of ISG expression was performed on mLNs and spleens harvested from WT SPF mice, WT SPF mice after 1 week of metronidazole (Met) treatment, or from Tlr4−/− SPF mice. Fold change gene expression in the mLN (E) or spleen (F) was calculated compared to WT SPF mice using the ΔΔCT method, with ActB as the reference gene and depicted as a heat map. Statistical analysis with (A) one-way ANOVA followed by Dunnett’s multiple comparisons test to WT, (B,D) two-way ANOVA followed by Dunnett’s multiple comparisons test to 0 ug/mL CTRL, and (C) unpaired t-test. (E,F) Details of statistical analyses can be found in Tables S3–4. ND=not detected, ns=not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
