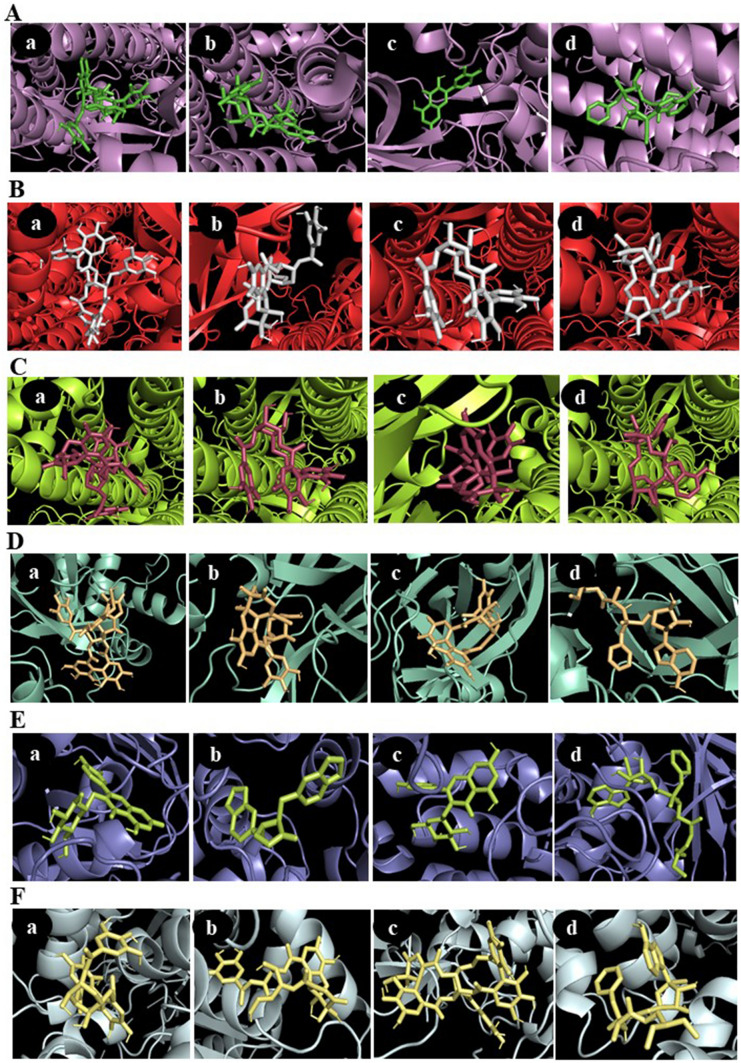
Fig. 2.
Output file obtained after docking analysis consists of top 9 binding poses with their respective binding affinity in kcal/mol. The ligand-binding pose showing highest binding affinity with least root mean square deviation (RMSD) was selected. The protein–ligand interaction in 3D structures were visualized in PyMOL. 3D visualization of interaction between SARS-CoV-2 proteins with representative phytochemicals. A S protein (6VSB) with a geraniin, b furosin, c quercetin-3-O-glucoside and d remdesivir (positive control). B S protein closed (6VXX) with a geraniin, b furosin, c corilagin and d Remdesivir (positive control) C S protein closed (6VYB) with a geraniin, b corilagin, c furosin and d remdesivir (positive control). D 3CLpro (6LU7) with a geraniin, b furosin, c corilagin and d remdesivir (positive control). E PLpro (4OVZ) with a quercetin-3-O-glucoside, b hinokinin, c astragalin and d remdesivir (positive control). F RdRp (6NUS) with a corilagin, b furosin, c geraniin and d remdesivir (positive control)