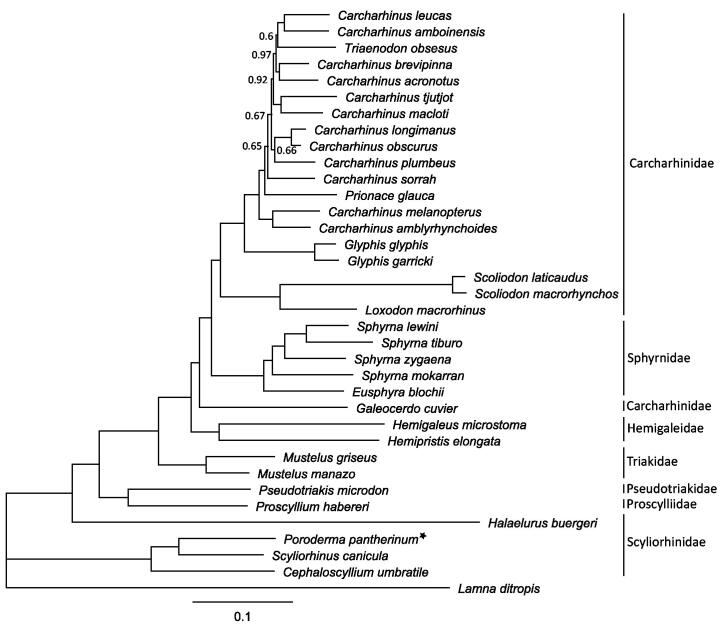
Figure 1.
Bayesian tree depicting the phylogenetic position of Poroderma pantherinum (posterior probability values only shown if below 1.0). Based on 35 mitochondrial genomes (excluding ND6 and the control region) of sharks from the order Carcharhiniformes, using Lamna ditropis (KF962053.1) as an outgroup. (Carcharhinus acronotus: NC_024055.1, Carcharhinus amblyrhynchoides: NC_023948.1, Carcharhinus amboinensis: NC_026696.1, Carcharhinus brevipinna: KM244770.1, Carcharhinus leucas: KF646785.1, Carcharhinus longimanus: NC_025520.1, Carcharhinus macloti: NC_024862.1, Carcharhinus melanopterus: NC_024284.1, Carcharhinus obscurus: NC_020611.1, Carcharhinus plumbeus: NC_024596.1, Carcharhinus sorrah: NC_023521.1, Carcharhinus tjutjot: KP091436.1, Cephaloscyllium umbratile: NC_029399.1, Eusphyra blochii: NC_031812.1, Galeocerdo cuvier: NC_022193.1, Glyphis garricki: KF646786.1, Glyphis glyphis: NC_021768.2, Halaelurus buergeri: NC_0311811.1, Hemigaleus microstoma: KT003687.1, Hemipristis elongata: KU508621.1, Loxodon macrorhinus: KT347599.1, Mustelus griseus: NC_023527.1, Mustelus manazo: NC_000890.1, Prionace glauca: NC_022819.1, Proscyllium habereri: KU721838.1, Pseudotriakis microdon: NC_022735.1, Scoliodon laticaudus: KP336547.1, Scoliodon macrorhynchos: NC_018052.1, Scyliorhinus canicula: NC_001950.1, Sphyrna lewini: NC_022679.1, Sphyrna mokarran: KY464952.1, Sphyrna tiburo: KM453976.1, Sphyrna zygaena: NC_025778.1 and Triaenodon obsesus: KJ748376.1).