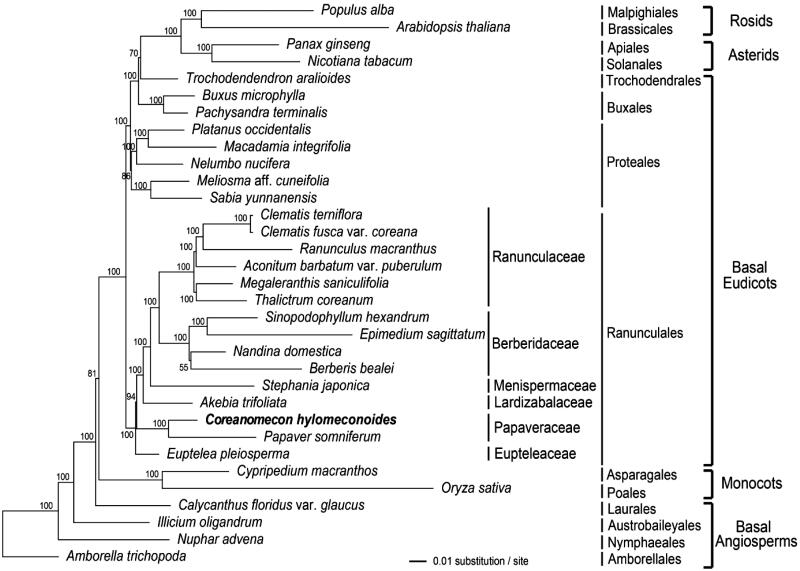
Figure 1.
Maximum likelihood (ML) tree based on 79 protein-coding and four rRNA genes from 33 plastid genomes as determined by RAxML (-ln L = -545811.818157). The numbers at each node indicate the ML bootstrap values. Genbank accession numbers of taxa are shown below, Aconitum barbatum var. puberulum (KC844054), Akebia trifoliata (NC_029427), Amborella trichopoda (NC_005086), Arabidopsis thaliana (NC_000932), Berberis bealei (NC_022457), Buxus microphylla (NC_009599), Calycanthus floridus var. glaucus (NC_004993), Clematis fusca var. coreana (KM652489), Clematis terniflora (NC028000), Coreanomecon hylomeconoides (KT274030, in this study), Cypripedium macranthos(NC_024421), Epimedium sagitatum (NC_029428), Eutelea pleiosperma (NC029429), Illicium oligandrum (NC_009600), Macadamia integrifolia (NC_025288), Megaleranthis saniculifolia (NC_012615), Meliosma aff. Cuneifolia (NC_029430), Nandina domestica (NC_008336), Nelumbo nucifera (NC_025339), Nicotiana tabacum (NC_001879), Nuphar advena (NC_008788), Oryza sativa (NC_001320), Pachysandra terminalis (NC_029433), Panax ginseng (NC_006290), Papavr somniferum (NC_029434), Platanus occidentalis (NC_008335), Populus alba(NC_008235), Ranunculus macranthus (NC_008796), Sabia yunnanensis (NC_029431), Sinopodophyllum hexandrum (NC_027732), Stephania japonica (NC_029432), Thalictrum coreanum (NC_026103), Trochodendron aralioides (NC_021426).