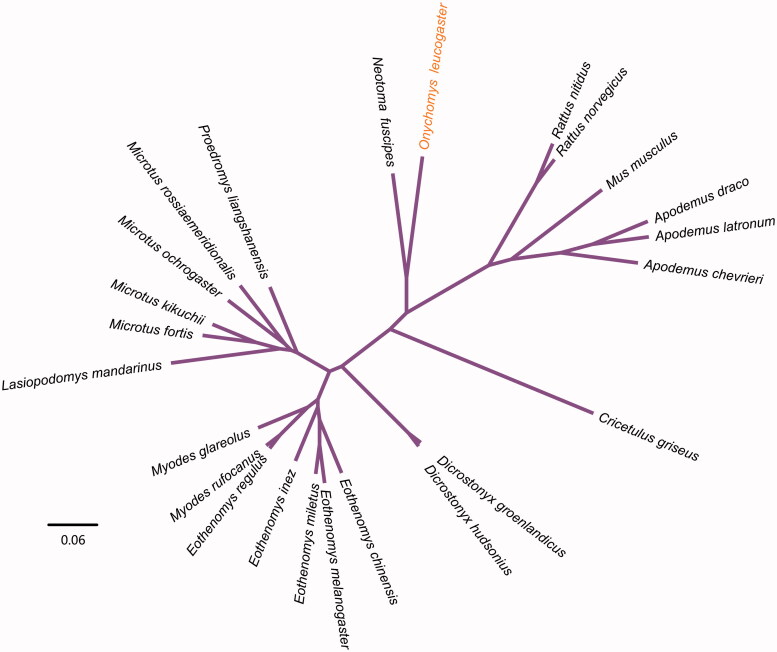
Figure 1.
Phylogenetic relationships among 24 rodent mt genoems. This tree was drawn without setting of a outgroup. All nodes exhibit full posterior probability (PP) and 100% RAxML supported bootstrap. The length of branch represents the divergence distance. Mitogenome accession numbers used in this phylogeny analysis: Apodemus chevrieri (HQ896683.1), Apodemus draco (HQ333255.1), Apodemus latronum (HQ333256.1), Cricetulus griseus (EU660217.1), Dicrostonyx groenlandicus (KX712239.1), Dicrostonyx hudsonius (KX683880.1), Eothenomys chinensis (FJ483847.1), Eothenomys inez (KU200225.1), Eothenomys melanogaster (KP997311.1), Eothenomys miletus (KX014874.1), Eothenomys regulus (JN629046.1), Lasiopodomys mandarinus (JX014233.1), Microtus fortis (JF261174.1), Microtus kikuchii (Microtus kikuchii), Microtus ochrogaster (KT166982.1), Microtus rossiaemeridionalis (DQ015676.1), Mus musculus (KF781655.1), Myodes glareolus (KF918859.1), Myodes rufocanus (KT725595.1), Neotoma fuscipes (KU745736.1), Proedromys liangshanensis (FJ463038.1), Rattus nitidus (KX058347.1), Rattus norvegicus (FJ919759.1).