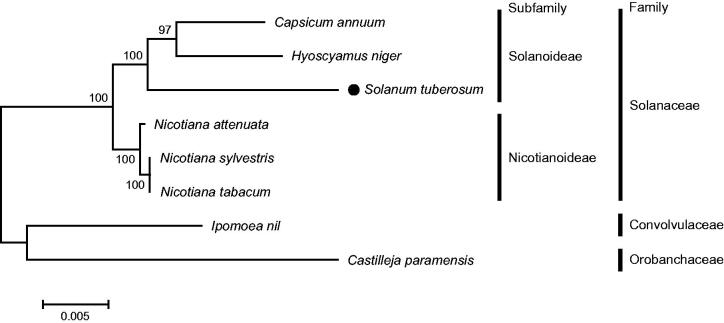
Figure 1.
Maximum-likelihood phylogenetic tree of Solanum tuberosum and related taxa based on mitochondrial genome sequences. Common protein-coding sequences in mitochondrial genome sequences were aligned using MAFFT (http://mafft.cbrc.jp/alignment/server/index.html) and used to generate maximum-likelihood phylogenetic tree by MEGA 6.0 (Tamura et al. 2013). Numbers on the nodes indicate bootstrap support values (>50%) from 1000 replicates. Scale bar represents the number of nucleotide substitution per site. Mitochondrial genome sequences used for this tree are Capsicum annuum, NC_024624; Castilleja paramensis, NC_031806; Hyoscyamus niger, NC_026515; Ipomoea nil, NC_031158; Nicotiana attenuata, MF579563; N. sylvestris, NC_029805; N. tabacum, NC_006581; Solanum tuberosum, MF989953–MF989957.