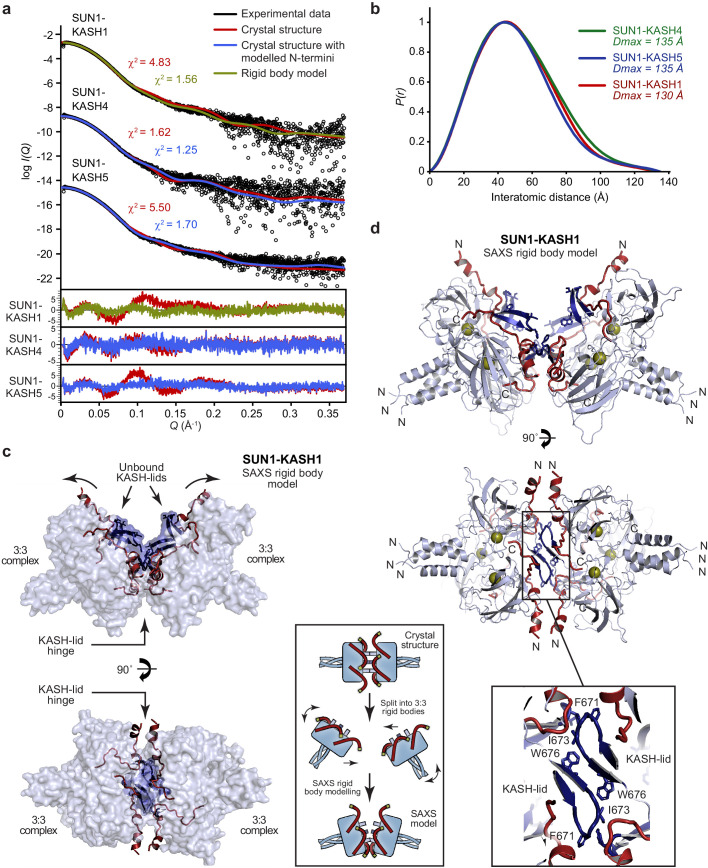
Figure 6. SEC-SAXS analysis of SUN-KASH 6:6 complexes.
(a) SAXS scattering curves of SUN1-KASH4, SUN1-KASH5, and SUN1-KASH1 overlaid with theoretical scattering curves of their crystal structures (red), crystal structures with KASH flexible N-termini modelled by CORAL (blue) and rigid body model of two 3:3 complexes (green). Residuals for each fit are shown (inset). Representative of more than three replicates using different protein preparations. (b) SAXS P(r) distributions showing maximum dimensions of 135 Å, 135 Å, and 130 Å, respectively. (c–d) SAXS rigid body model of SUN1-KASH1 shown as (c) surface and (d) cartoon representation, in which two constituent 3:3 complexes from its crystal structure were assigned as rigid bodies, with the 6:6 assembly generated by fitting to experimental SAXS data of solution SUN1-KASH1 (χ2 = 1.56). The inlet schematic illustrates the SAXS rigid body modelling procedure in which the crystal structure was split into its constituent 3:3 complexes, which were rotated as rigid bodies in three dimensions and allowed to interact, whilst fitted against experimental SAXS data. (d) The cartoon representation highlights structural details of the predicted KASH-lid interface, including the presence of unbound KASH-lids, and the close approximation of opposing KASH-lids, which achieve an asymmetric positioning of the N-termini of KASH domains in locations and orientations compatible with their upstream sequences crossing the outer nuclear membrane.
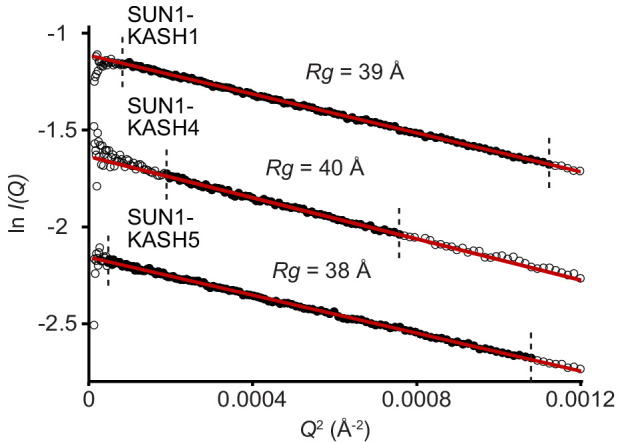
Figure 6—figure supplement 1. SAXS analysis of SUN-KASH complexes.