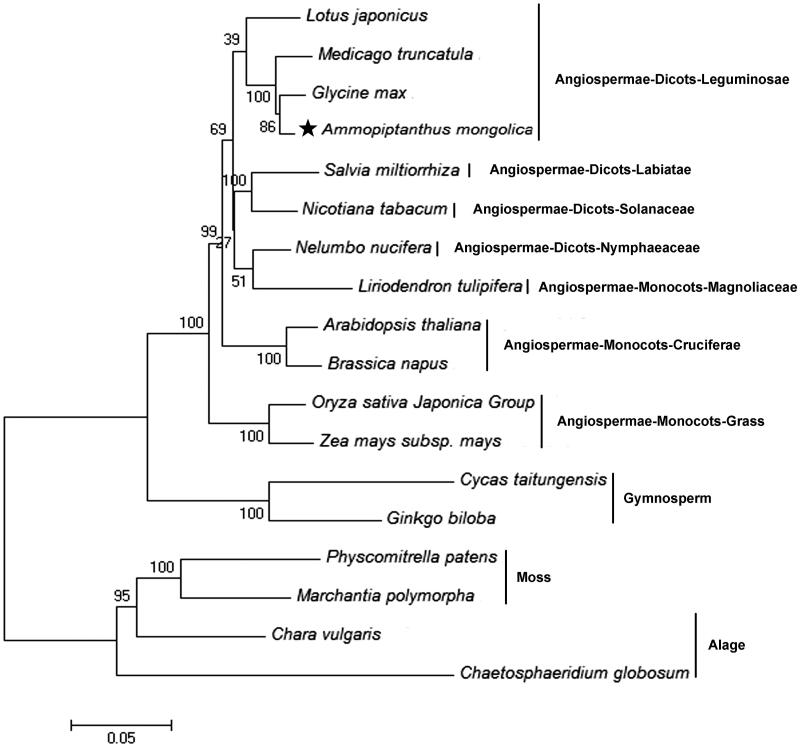
Figure 1.
Phylogenetic tree of Ammopiptanthus mongolicus and 17 other plant mitochondrial genomes. Neighbour-joining method of 17 amino acid sequences were used, including atp8, atp9, cob, cox1, cox2, cox3, nad1, nad3, nad4L, nad5 and nad6. All the sequences could be currently available in the GenBank database: Lotus japonicus (NC_016743), Medicago truncatula (NC_029641), Glycine max (NC_020455), Salvia miltiorrhiza (NC_023209), Nicotiana tabacum (NC_006581), Nelumbo nucifera (NC_030753), Liriodendron tulipifera (NC_021152), Arabidopsis thaliana (NC_001284), Brassica napus (NC_008285), Oryza sativa Japonica Group (NC_011033), Zea mays subsp. mays (NC_007982), Cycas taitungensis (NC_010303), Ginkgo biloba (NC_027976), Physcomitrella patens (NC_007945), Marhantia polynorpha (NC_001660), Chara vulgaris (NC_005255), and Chaetosphaeridium globosum (NC_004118); Chaetosphaeridium globosum was used as the outgroup. Numbers above each node represent bootstrap values from 1000 replicates. Black star indicate Ammopiptanthus mongolicus.