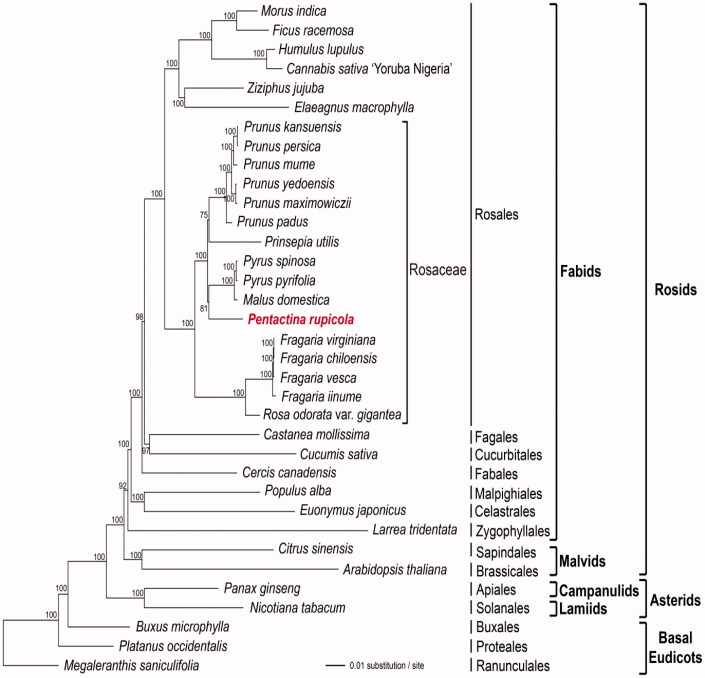
Figure 1.
ML tree based on 82 protein-coding and rRNA genes from 35 plastid genomes as determined by RAxML (−ln L = −441,234.528222). The numbers at each node indicate the ML bootstrap values. Genbank accession numbers of used taxa are shown below, Arabidopsis thaliana (NC_000932), Buxus microphylla (NC_009599), Canabis sativa ‘Yoruba Nigeria’ (NC_027223), Castanea mollissima (NC_014674), Cercis canadensis (KF856619), Citrus sinensis (NC_008334), Cucumis sativus (NC_007144), Elaeagnus macrophylla (NC_028066), Euonymus japonicus (NC_028067), Ficus racemosa (NC_028185), Fragaria chiloensis (NC_019601), F. iinumae (NC_024258), F. vesca (NC_015206), F. virginiana (NC_019602), Humulus lupulus (NC_028032), Larrea tridentata (NC_028023), Malus domestica (Genome Database for Rosaceae), Megaleranthis saniculifolia (NC_012615), M. indica (NC_008359), N. tabacum (NC_001879), P. ginseng (NC_006290), P. rupicola (NC_016921, in this study), Platanus occidentalis (NC_008335), Populus alba (NC_008235), Prinsepia utilis (NC_021455), Prunus kansuensis (NC_023956), P. maximowiczii (NC_026981), P. mume (NC_023798), P. padus (NC_026982), P. persica (NC_014697), P. yedoensis (NC_026980), P. pyrifolia (NC_015996), P. spinosa (NC_023130), Rosa odorata var.gigantea (KF753637), and Ziziphus jujuba (NC_030299).