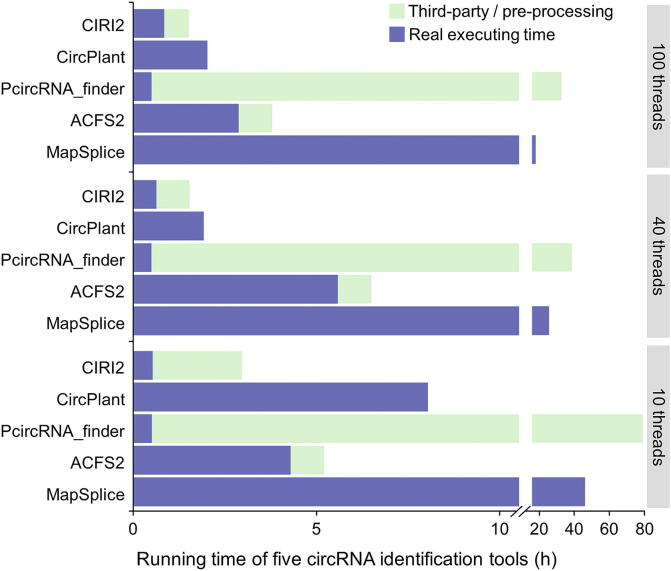
Figure 2.
Performance comparison of five circRNA detection tools
Five circRNA identification tools are tested on a same RNA-seq dataset. For each tool, the total executing time was separated into two parts: time for the dependencies (third-party or pre-processing programs needed by the tool) and time for the main algorithm (real executing time). The results are represented by stacked bars with different colors. MapSplice and CircPlant did not have executing time for third-party / pre-processing procedures, as their dependencies are integrated into their main routines. For procedures with parallel processing, only the longest executing time was counted.