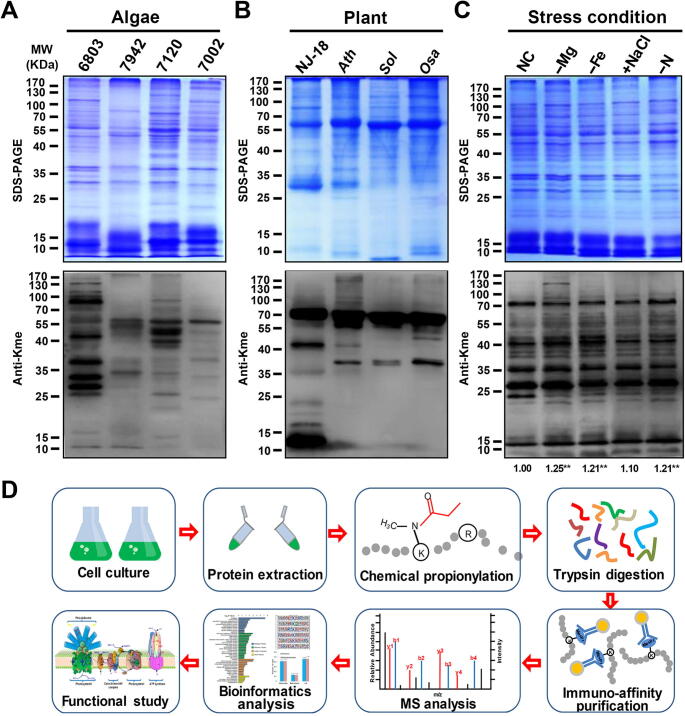
Figure 1.
Profiling lysine methylation in photosynthetic organisms
A. Detection of lysine methylation in Synechocystis sp. PCC 6803 (6803), Synechococcus sp. PCC 7942 (7942), Anabaena sp. PCC 7120 (7120), and Synechococcus sp. PCC 7002 (7002) using 20 μg of proteins and anti-monomethyllysine (anti-Kme) antibody. B. Detection of lysine methylation in Chlorella sp. NJ-18 (NJ-18), Arabidopsis thaliana (Ath), Spinacia oleracea (Sol), and Oryza sativa (Osa) using 10 μg of proteins and anti-Kme antibody. C. Profiling of lysine methylation in Synechocystis sp. PCC 6803 under various stress conditions. Proteins (20 μg) were extracted from the cells cultured under normal condition (NC), magnesium deficiency (−Mg), ferric deficiency (−Fe), sodium chloride enrichment (+NaCl), or nitrogen deficiency (−N). A densitometry analysis was performed using ImageJ software. Samples were standardized through comparisons with Coomassie-blue-stained gel. Data represent results from three independent experiments. The level of statistical significance was determined using two-sample Student’s t-test (**, P < 0.01). D. Flowchart illustrating the experimental procedure for lysine monomethylation proteomics.