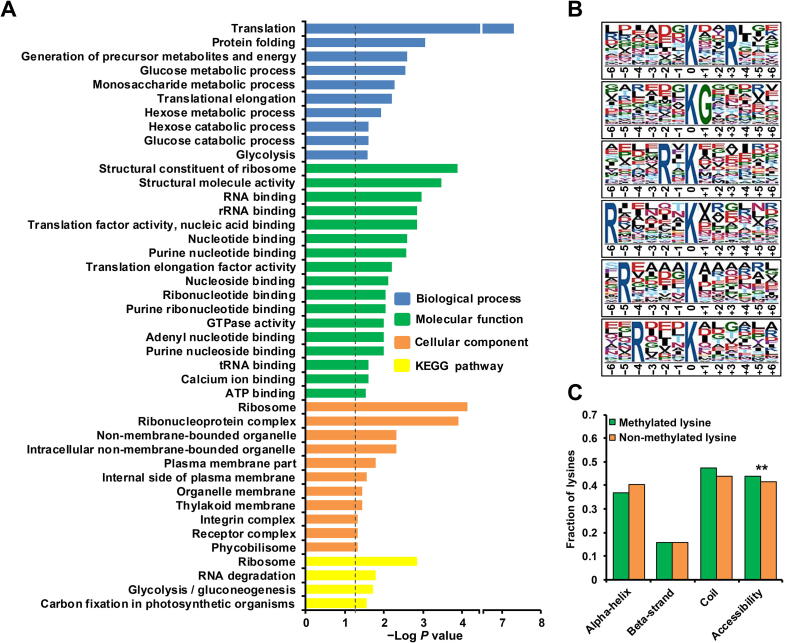
Figure 2.
Enrichment analysis of monomethylated proteins and characterization of lysine monomethylation sites
A. Bar graph showing the enrichment of monomethylated proteins for Gene Ontology (GO) categories (including biological processes, molecular functions, and cellular components) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The enrichment of GO categories and KEGG pathways was carried out using DAVID bioinformatics tools (P < 0.05). B. Sequence motifs for all lysine monomethylation sites. The significant motifs were identified by Motif-X software with a corresponding P < 0.000001. C. Distributions of methylated and non-methylated lysines in structured and unstructured regions of the proteins. Statistical significance was determined by two-sample Student’s t-test (**, P < 0.01).