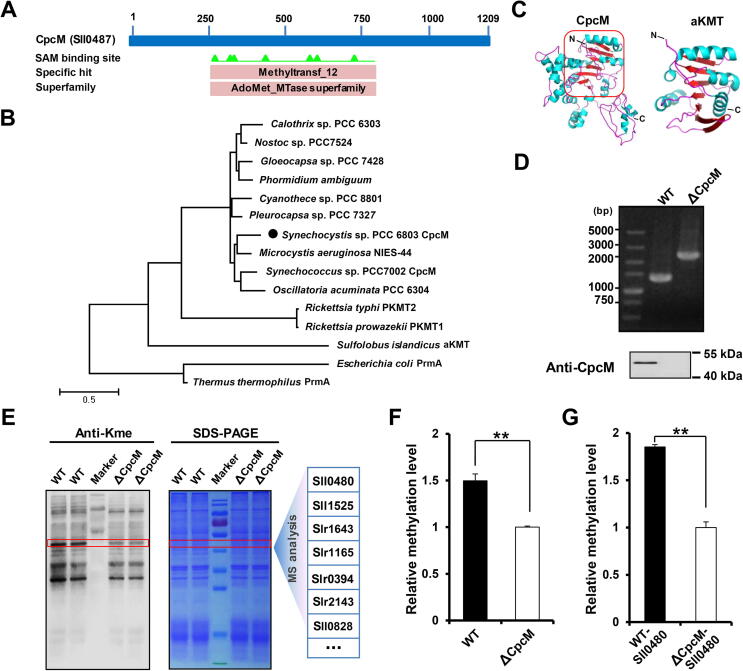
Figure 4.
CpcM has a lysine methyltransferase activity in Synechocystis
A. CpcM was predicted to be a potential SAM-dependent methyltransferase. B. Evolutionary conservation of lysine methyltransferases. The maximum likelihood phylogenetic tree was built using MEGA 5.0.5 software. C. Homology model of CpcM and its ortholog aKMT from Sulfolobus islandicus. D. Verification of the mutant lacking CpcM by PCR and Western blot. E. Identification of lysine methylated substrates. Three pairs of bands from the total cell lysates of WT and ΔCpcM in Coomassie-blue-stained gel, which exhibited varying levels of lysine methylation by Western blot analysis, were cut from three replicate gels and trypsin-digested in-gel, followed by LC-MS/MS analysis. F. Quantitative analysis of the corresponding methylated proteins from the bands in (E). The intensities of modified peptides were normalized using the identified protein abundances determined by MaxQuant software. G. Quantitative analysis of methylated Sll0480 from the bands in (E). Data are presented as means ± SD from three independent experiments. A two-sample Student’s t-test was performed to determine the level of statistical significance (**, P < 0.01).