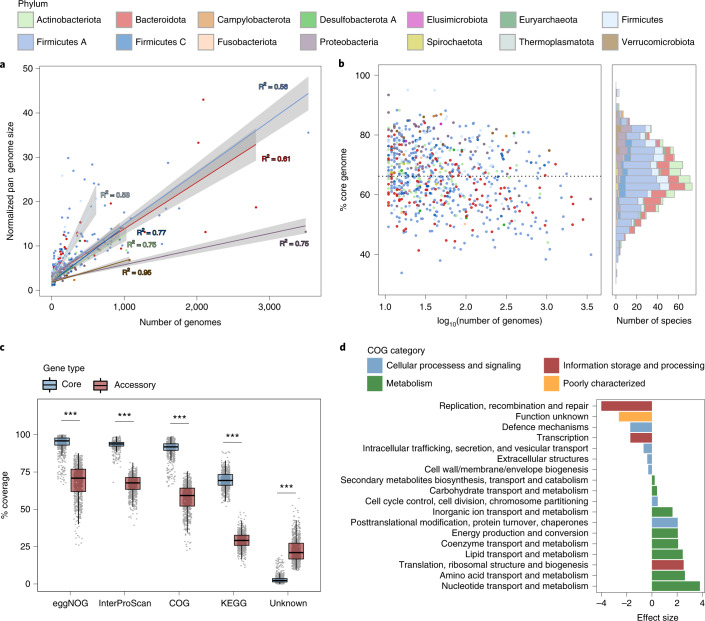
Fig. 5. Pan-genome diversity patterns within the gut microbiome.
a, Normalized pan-genome size as a function of the number of conspecific genomes. Regression curves were generated for each phylum, with the corresponding coefficients of determination indicated next to each curve and the shaded regions representing the 95% confidence level intervals. The following correspond to the number of species considered for each phylum: Actinobacteriota, n = 66; Bacteroidota, n = 122; Firmicutes, n = 90; Firmicutes A, n = 325; Firmicutes C, n = 44; Proteobacteria, n = 65; Verrucomicrobiota, n = 13. b, Fraction of the core genome for each species according to the number of conspecific genomes (left) and as a histogram (right), colored by phylum. The horizontal dashed line represents the median value across all species. c, Proportion of core and accessory genes (n = 781 species) classified with various annotation schemes, alongside the percentage of genes lacking any functional annotation. Box lengths represent the IQR of the data, and whiskers extend to the lowest and highest values within 1.5 times the IQR from the first and third quartiles, respectively. A two-tailed Wilcoxon rank-sum test was performed to compare the classification between the core and accessory genes (***P < 0.001). d, Comparison of the functional categories assigned to the core (n = 1,236,880) and accessory (n = 4,785,975) genes. Only statistically significant (adjusted P < 0.05) differences are shown. Significance was calculated with a two-tailed Wilcoxon rank-sum test and further adjusted for multiple comparisons using the Benjamini–Hochberg correction. A positive effect size (Cohen’s d) indicates over-representation in the core genes.