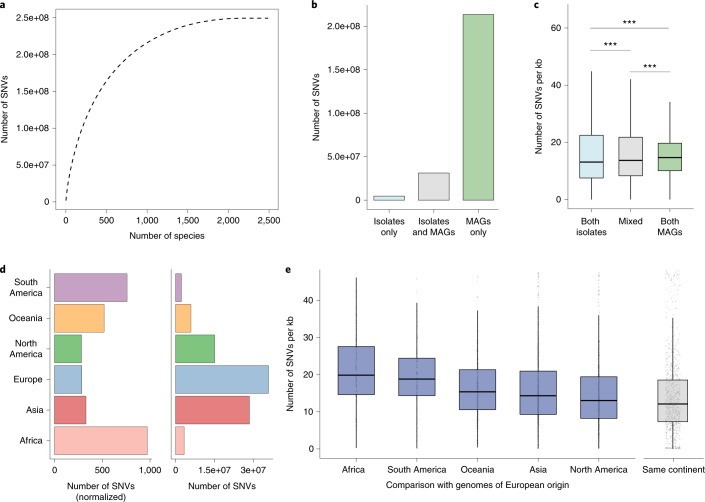
Fig. 6. Analysis of intraspecies single-nucleotide variation.
a, Total number of SNVs detected as a function of the number of species. The cumulative distribution was calculated after ordering the species by decreasing number of SNVs. b, Number of SNVs detected only in isolate genomes or MAGs, or in both. c, Pairwise SNV density analysis of genomes of the same or different type (isolates, n = 808,331 comparisons; mixed, n = 1,575,895 comparisons; MAGs, n = 26,899,457 comparisons). A two-tailed Wilcoxon rank-sum test was performed to assess statistical significance and further adjusted for multiple comparisons using the Benjamini–Hochberg correction (***P < 0.001). d, Left, the number of exclusive SNVs normalized by the number of genomes per continent. Right, the number of SNVs exclusively detected in genomes from each continent. e, Pairwise SNV density analysis between genomes from Europe, the largest genome subset, and other continents. The median SNV density was calculated per species, and the distribution is shown for all species (Africa, n = 188; Asia, n = 746; North America, n = 688; Oceania, n = 35; South America, n = 151). Comparison of genomes recovered from the same continent (n = 908 species) was used as a reference. The SNV density between genomes from the same continent is significantly lower (adjusted P < 0.05) than that calculated for genomes from different continents. In c and e, box lengths represent the IQR of the data, with whiskers depicting the lowest and highest values within 1.5 times the IQR of the first and third quartiles, respectively.