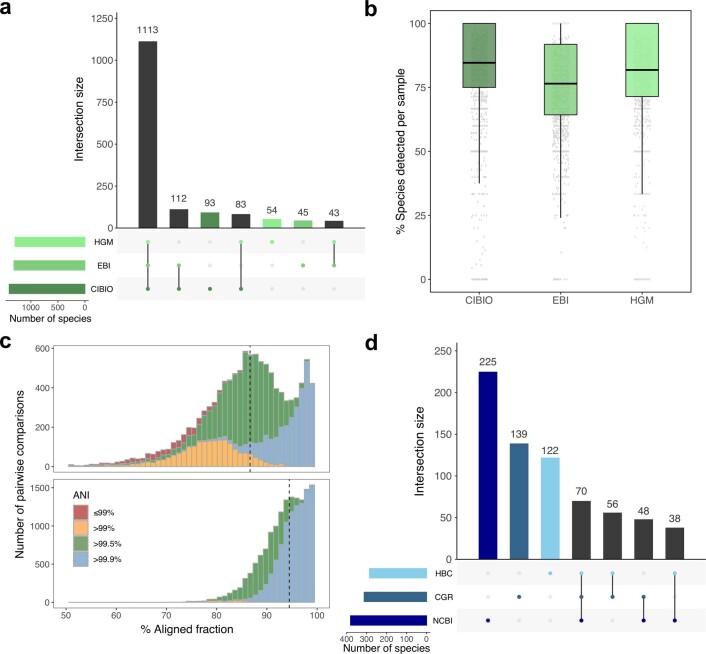
Extended Data Fig. 3. Species overlap across study sets.
a, Number of species found across the three metagenome-assembled genome sets, ordered by their level of overlap. Only those genomes recovered from the 1,554 metagenomic samples used by all three studies were considered in this analysis. b, Distribution of the proportion of species recovered per sample (n = 1,554) in each study set out of all species recovered across all three studies in the same samples. Box lengths represent the IQR of the data, and the whiskers the lowest and highest values within 1.5 times the IQR from the first and third quartiles, respectively. c, Estimated aligned fractions and average nucleotide identities (ANI) between conspecific genomes obtained in the same sample but in different MAG studies. Results for medium-quality genomes are illustrated in the top panel, whereas those for near complete (≥90% completeness) genomes are represented in the lower panel. Vertical dashed lines denote the median values. d, Number of species identified in three culture-based studies and their degree of overlap. The NCBI study set consists mainly of genomes from the Human Microbiome Project (HMP).