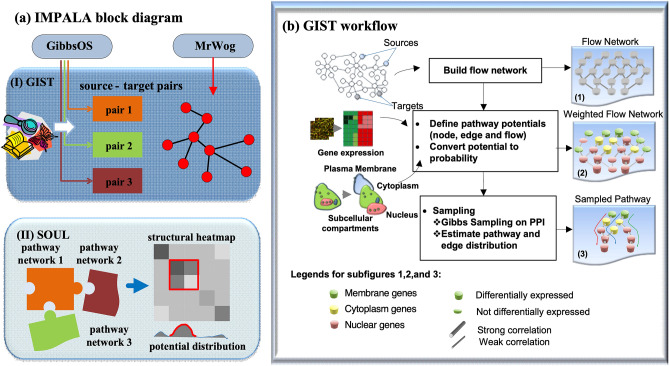
Figure 1.
IMPALA block diagram and GIST workflow. (a) Key transcription factors and the candidate pathway landscape are identified using GibbsOS and MrWOG to pre-process gene expression and protein–protein interaction data (HPRD database). Then, IMPALA integrates gene expression and candidate pathways to identify aberrant signal pathway transduction using GIST (Gibbs sampler to Infer Signal Transduction) and pathway modules using SOUL (Structural Organization to Uncover pathway Landscape). (b) GIST integrates gene (node), gene–gene interaction (edge) and network flow potentials to build a weighted and directed Bayesian network and infers signaling directions between genes using Gibbs Sampling.