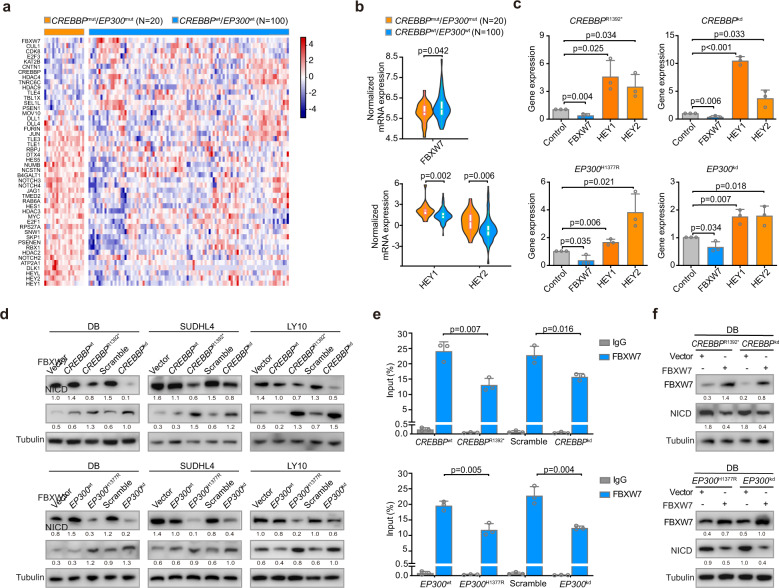
Fig. 4.
CREBBP/EP300 mutations inhibited FBXW7 and activated the NOTCH signaling pathway. a Heatmap of genes associated with the NOTCH signaling pathway in CREBBPmut/EP300mut patients, as compared to CREBBPwt/EP300wt patients. b Normalized mRNA expression of FBXW7, HEY1 and HEY2 in tumor samples of diffuse large B-cell lymphoma (DLBCL) patients with or without CREBBP/EP300 mutations as revealed by RNA sequencing data. c Relative gene expression of FBXW7, HEY1, and HEY2 in CREBBPR1392*, CREBBPkd, EP300H1377R, and EP300kd DB cells, as compared to CREBBPwt, EP300wt or scramble DB cells by quantitative real-time PCR (RT-PCR). Data are presented as the mean ± SD (N = 3). d Protein expression of FBXW7 and NICD detected in vector, CREBBPwt, CREBBPR1392*, scramble, CREBBPkd DB, SUDHL4, and LY10 cells, and in vector, EP300wt, EP300H1377R, scramble, EP300kd DB, SUDHL4 and LY10 cells by western blot. The same lysates were used as in Fig. 3b. Tubulin was used as a loading control. The FBXW7/Tubulin and NICD/tubulin ratio were shown. e Occupancies of H3K27ac in the proximal promoter areas of FBXW7 in CREBBPwt, CREBBPR1392*, scramble, CREBBPkd DB cells, and in EP300wt, EP300H1377R, scramble, EP300kd DB cells by chromatin immunoprecipitation (ChIP) assay. Data are presented as the mean ± SD (N = 3). f Expression of NICD in CREBBPR1392* and CREBBPkd DB cells, as well as EP300H1377R and EP300kd DB cells with or without reintroduction of FBXW7 protein by western blot