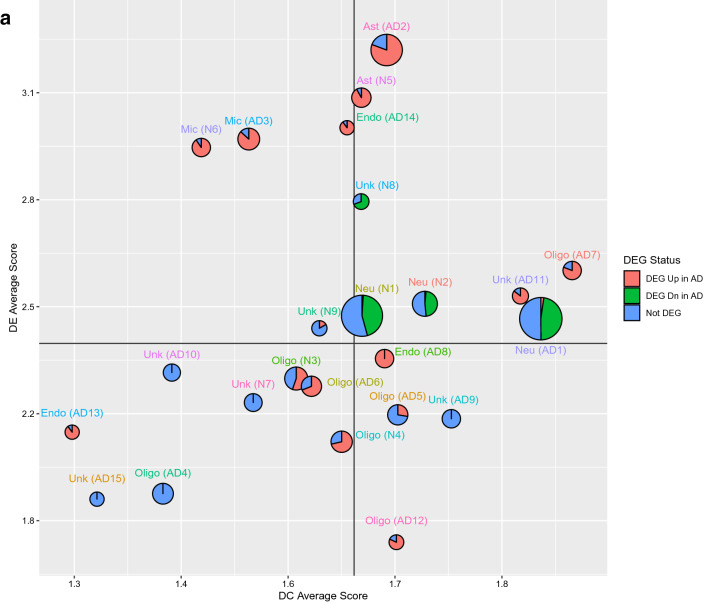
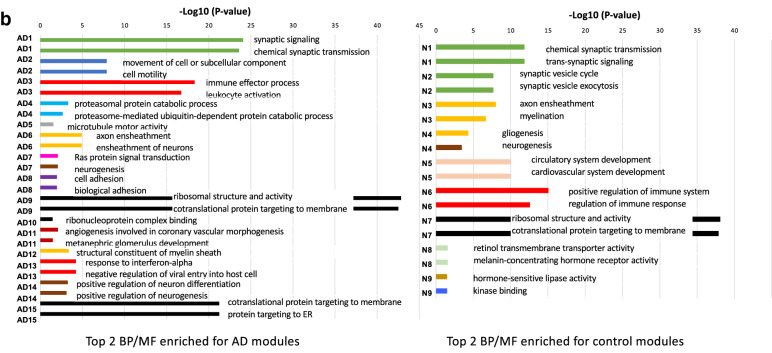
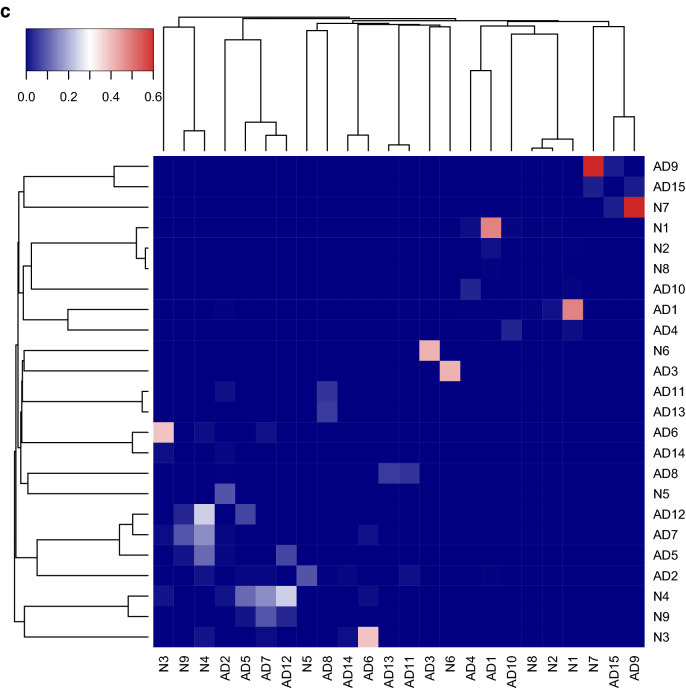
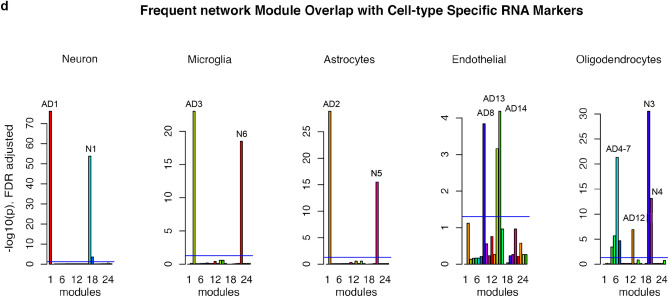
Figure 1.
FGCN modules from AD and control samples classified according to differential expression and co-expression levels. (a) Differential expression average scores (DE) vs. differential co-expression average scores (DC) of 24 frequent gene co-expression network modules (module names in parenthesis), the majority of which are enriched with specific biological functions as shown in panel b. Most of the modules are also enriched for specific cell-type markers: Neu, neurons; Ast, astrocytes; Mic, microglia; Endo, endothelia; Oligo, oligodendrocytes; Unk, modules not enriched with cell type-specific markers. The pie charts indicate the proportions of differentially expressed genes (up or down in AD vs. control) in the modules. (b) The top two enriched biological functions/processes/pathways in each module identified using ToppGene. The colors of the bars are indictive of different biofunctional terms. (a). (c) A heatmap of the degree of overlap between the genes in each pair of modules mined from the AD and control samples. The modules were rearranged by their similarities and then plotted according to the Jaccard Index values. (d) Cell-type enrichment for each module was assessed by cross-referencing the module genes against cell-type signatures of neurons, microglia, astrocytes, oligodendrocytes, and endothelial cells. The x-axis shows the different modules. The 24 FGCN modules are numbered as: 1–15 correspond to modules AD1 to AD15; 16–24 correspond to modules N1 to N9. The significance of the cell-type enrichment in the modules was measured by Fisher’s exact test and corrected for multiple comparisons by the Benjamini and Hochberg procedure. Horizonal blue lines indicate the threshold of false discovery rate 0.05. There is no color match between panels (a,d).