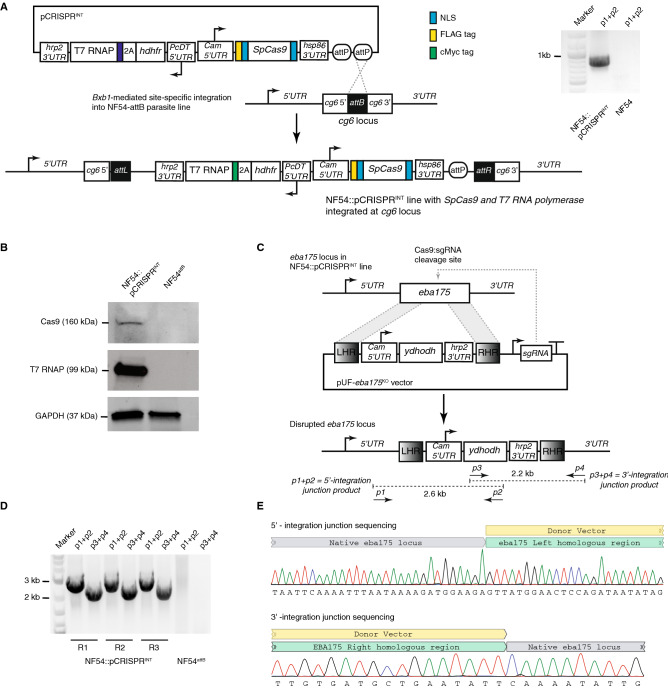
Figure 4.
Validation of a pCRISPR plasmid reagent for CRISPR/Cas9-mediated genome engineering via transient or constitutive SpCas9 expression. (A) The pCRISPR plasmid allows constitutive expression of SpCas9 from the CAM 5′-UTR/hsp86 3′-UTR transcription unit. Through use of a T2A “skip peptide”, both the human dihydrofolate reductase (hdhfr) selectable marker and T7 RNA polymerase for sgRNA transcription are expressed from the PcDT 5′-UTR/hrp2 3′-UTR transcription unit. The 2 × attP sites allow for Bxb1-mediated integration into a chromosomal attB site previously engineered into a host parasite strain. Successful integration of pCRISPR plasmid at the cg6 locus in an NF54::pCRISPR parasite line was confirmed by PCR analysis. (B) Western blot analysis to detect expression of SpCas9 (anti-FLAG) and T7 RNA polymerase (anti-Myc) proteins by the NF54::pCRISPR parasite. (C) Schematic of the donor vector used to disrupt the dispensable eba175 gene, and configuration of the eba175 locus in the NF54::pCRISPR line pre- and post-editing. The donor vector contained left and right homologous regions from eba175 flanking a selectable ydhodh expression cassette, and a T7 RNAP-transcribed cassette for expression of the sgRNA targeting eba175. (D) PCR analysis to detect the 5′- and 3′-integration junctions in edited lines using the primer pairs p1 + p2 and p3 + p4, respectively. (E) Sanger sequencing data for the 5′- and 3′-integration junction PCR products obtained from the knockout transfection replicate, R3.