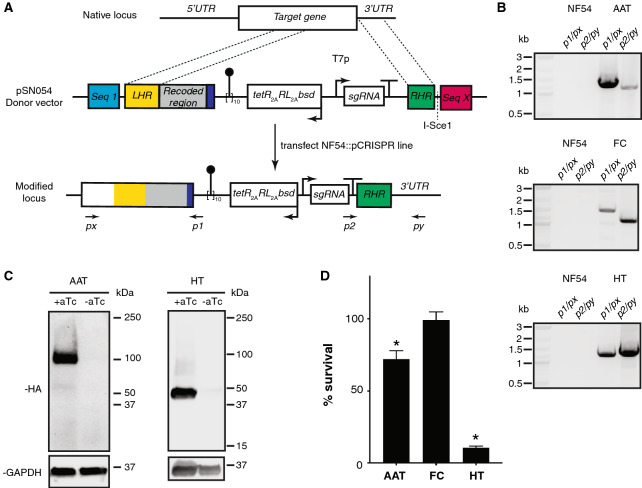
Figure 7.
Manipulating the 3′-UTRs of native loci using linear pSN054 donor vectors and CRISPR/Cas9 genome editing. (A) Generalized schematic of CRISPR/Cas9-mediated editing of a target locus to install C-terminal 2x-HA epitope tags, a 10 × TetR aptamer array, and a TetR-DOZI, Renilla luciferase and Blasticidin S deaminase expression cassette. Donor vectors were transformed into the NF54::pCRISPR parasite line to modify targeted loci. (B) gDNA extracted from transgenic parasites analyzed by PCR demonstrated formation of the expected 5′- and 3′- integration junctions at each targeted locus. Expected junctional PCR product sizes (5′, 3′): AAT, putative amino acid transporter (1.4 kb, 1.1 kb); FC, ferrochelatase (1.5 kb, 1.2 kb); and HT, hexose transporter (1.3 kb, 1.4 kb). Marker = 1 kb Plus DNA ladder (New England Biolabs). Full gel images are included in Supplementary Fig. 6A–C. (C) Western blot analysis of target protein expression under ± aTc conditions. Expected molecular weights of the 2 × HA-tagged proteins are AAT = 137.6 kDa and HT = 60.5 kDa. No HA-tagged FC protein was detected, even after multiple attempts. Note: AAT and HT migrate faster than expected based on their molecular weights; however, N- and C-terminal tagged HT migrate identically (Fig. 5). Full gel images are included in Supplementary Fig. 6D–E. (D) Normalized Renilla luciferase levels for parasites grown in 0 nM or 50 nM aTc for 72 h. Data represent the mean of n = 3 ± s.e.m. * p ≤ 0.05 by Student’s t-test.