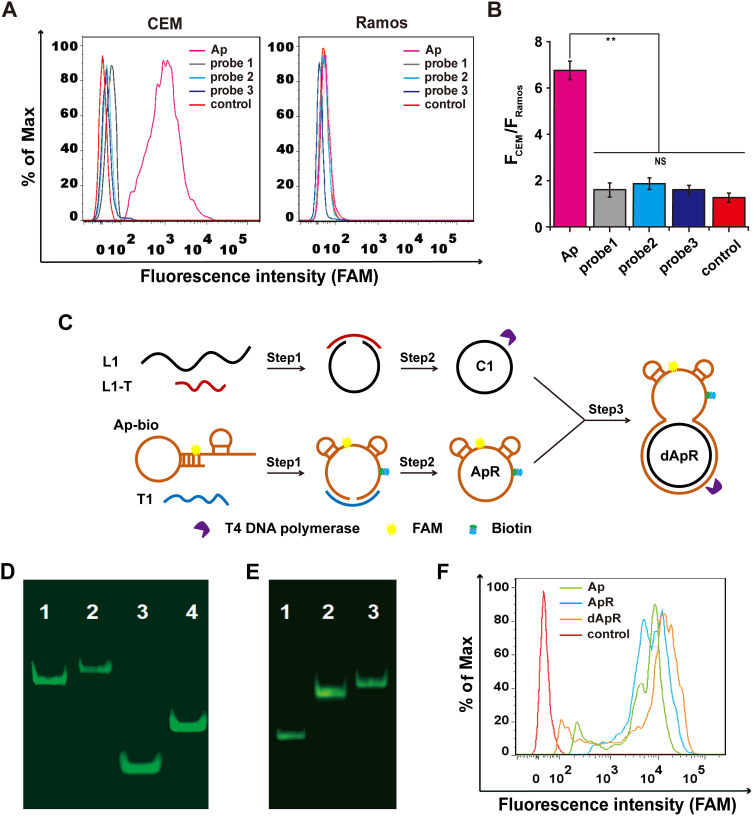
Figure 1.
Design and synthesis of the double-strand (ds) circular aptamer (dApR) and their biostability and targeting functionality. (A) Flow cytometry assays proved that Aptamer (Ap) targeted CEM (target cells), but not the control Ramos (non-target cells), while the control probes (probe1, probe2 and probe3) did not bind to CEM and Ramos cells. (B) Quantitative fluorescence ratios of the target CEM cells to the control Ramos cells for binding Ap. (C) The procedure for preparing dApR. L1 hybridized with L1-T, while biotinylated Ap (Ap-bio) hybridized with T1, and then single strand circular DNAs (C1 and ApR) were obtained by ligation. ApR and C1 hybridized with each other to turn into the double strand circular aptamer, namely, dApR. (D) The successful synthesis of dApR was confirmed by comparison of difference in mobility between Ap and dApR, or T1 and C1 in PAGE gel. Lane 1: Ap, Lane 2: dApR, Lane 3: T1, Lane 4: C1. (E) The biostability of ApR and dApR was evaluated by PAGE gel analysis after their incubation with T4 DNA polymerase (5 μM) for 2 h, while Ap (Lane 1) degrade, but not ApR (Lane 2) and dApR (Lane 3). (F) Flow cytometry data showed that dApR bound to CEM cells more specifically than Ap and ApR did, indicating the greater binding efficiency. NS and ** respectively indicates no statistical difference and P < 0.01.