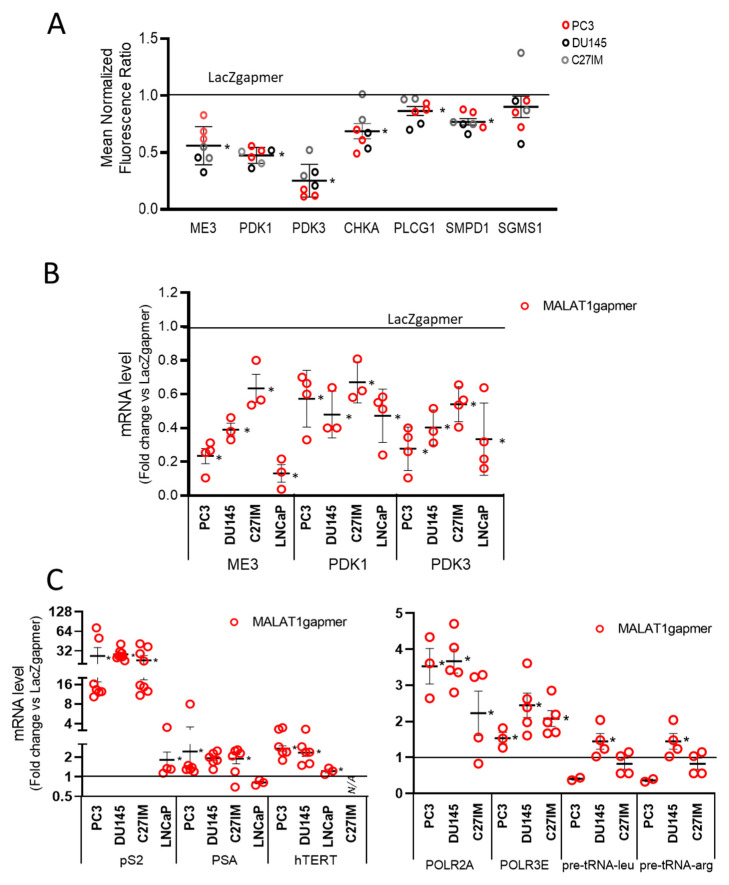
Figure 3.
Validation of array data in PCa cells by qRT-PCR. (A) Differential expression of genes as emerging from transcriptomic analysis before/after MALAT1 depletion in C27IM, DU145, and PC3 cells. Transcript levels of ME3, PDK3, PDK1, and PCho-related genes (choline kinase A (CHKA), Phospholipase C Gamma 1 (PLGC1), sphingomyelin phosphodiesterase 1 (SMPD1), and sphingomyelin synthase 1 (SGMS1)) are expressed as fold change vs. LacZgapmer (mean ± SEM are showed). (B,C) mRNA expression level of those genes emerging as differentially modulated were validated in different PCa cells (PC3, DU145, C27IM, and LNCaP) transfected with specific gapmer for MALAT1 or LacZ. Results are plotted as fold induction vs. LacZgapmer (placed to 1 and depicted as a black line). Individual values (n = 3–8) with mean ± SEM are showed. Non-parametric paired two-tailed Student’s t-test determined statistical significance. * p ≤ 0.05 MALAT1gapmer vs. LacZgapmer. * p < 0.05 vs. gapmerLacZ.