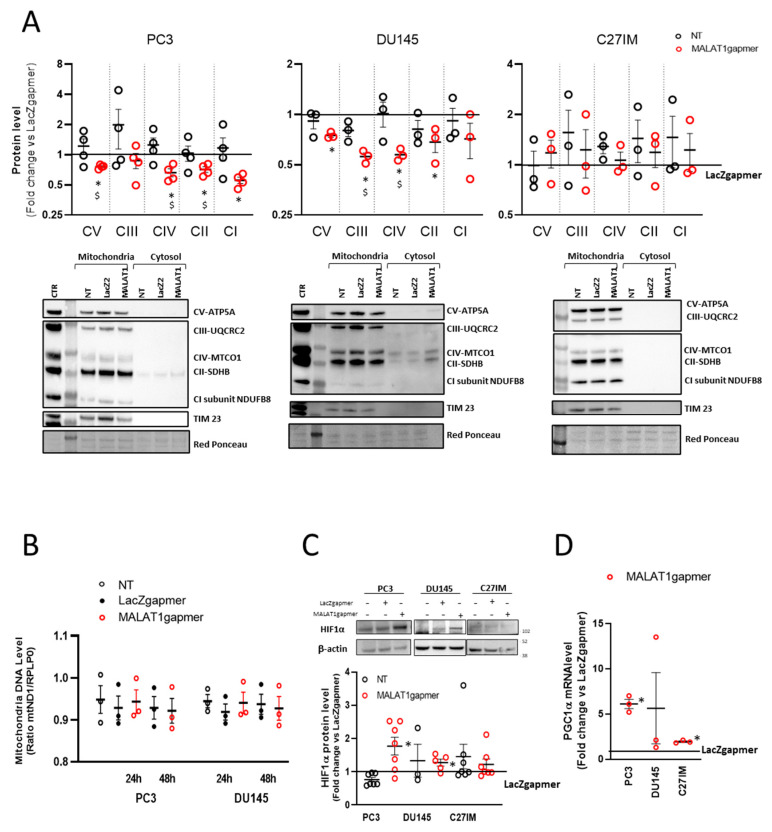
Figure 4.
Metabolic alteration after MALAT1 depletion. (A) Western blotting analysis of OXPHOS complex subunit expression determined by using total OXPHOS antibody cocktail in PC3 (left), DU145 (middle), and C27IM (right) cells’ mitochondrial fractions upon MALAT1 depletion. Upper panels: Densitometric analysis plotted as fold change vs. LacZgapmer after normalization to loading control red ponceau. Lower panel: Representative western blotting experiments. Uncropped Western Blots of Figure 4A are available in Figure S9 (B) qRT quantification of mitochondrial DNA level in PC3 and DU145 cells at 24 h or 48 h after transfection with specific (MALAT1), control (LacZ), or no (NT) gapmer. Data are expressed as ratio mtND1 vs. RPLP0 level. (C) Representative western blot and densitometry analysis for HIF1α assessed in 3 different PCa cells (PC3, DU145, and C27IM) not transfected or transfected with MALAT1 or LacZgapmers. β-actin was used as a loading control. Uncropped Western Blots of Figure 4C are available in Figure S10 (D) qRT expression of PGC1alpha mRNA in different PCa cells (PC3, DU145, and C27IM) interfered with MALAT1 or LacZgapmers. Results are plotted as fold induction vs. LacZgapmer taken as reference value 1 (straight black line). Individual values (n = 3–8) with mean ± SEM are shown. Non-parametric paired two-tailed Student’s t-test determined statistical significance. * p ≤ 0.05 MALAT1gapmer vs. LacZgapmer. $ p ≤ 0.05 MALAT1gapmer vs. NT.