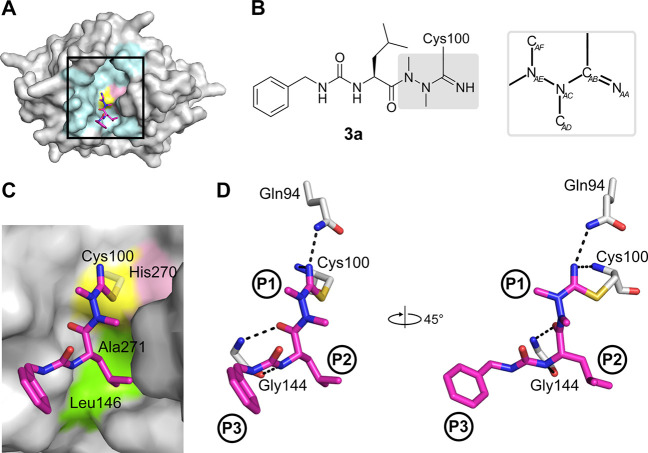
Figure 4.
Binding mode of the azadipeptide nitrile inhibitor 3a in the SmCB1 active site. (A) Overall crystal structure of the SmCB1–3a complex in surface (enzyme) and stick (inhibitor) representation. In the SmCB1 active site (boxed), the catalytic residues Cys100 (yellow) and His270 (pink) and major subsite residues (cyan) are highlighted. (B) Chemical structure of 3a forming a covalent bond with the S atom of the catalytic Cys100. The azanitrile warhead is boxed in gray, and atom labeling is indicated (hydrogen atoms are omitted). (C) Zoomed view of (A) showing the active site residues that form nonpolar interactions (green) with 3a (in stick representation with carbon atoms in magenta); the catalytic residues are also indicated. (D) The P1 to P3 positions of the inhibitor bind the corresponding S1 to S3 subsites of the SmCB1 active site. Dashed lines indicate hydrogen bonds formed between SmCB1 residues (gray) and 3a (magenta); heteroatoms have standard color coding (O, red; N, blue; S, yellow). Coordinates are deposited under PDB code 6YI7.