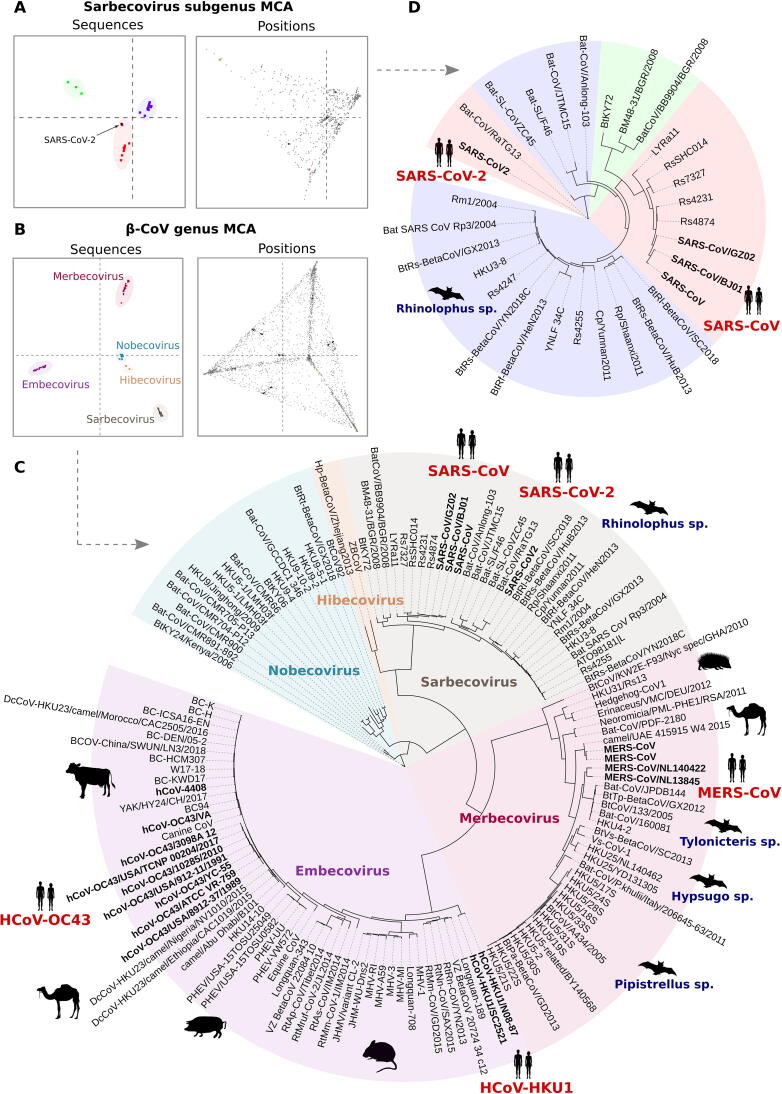
Fig. 1.
Results of the S3Det MCA analysis based on the full β-CoVs family and Sarbecovirus subgroup. (A-B) Results of the S3Det MCA analysis showing the subfamily segregation and associated amino acid positions obtained for the Sarbecovirus subgroup and for the full β-CoV family, respectively. (C) Phylogenetic tree obtained for the complete β-CoV spike protein family. S3Det subfamilies are shown in different colors and reflect the phylogenetic classification of Betacoronavirus into five subgenera. (D) Phylogenetic tree obtained for the Sarbecovirus subgroup. S3Det clusters are highlighted in red, green and blue. Both SARS-CoV-2 and RaTG13 are clustered together with SARS-CoV and other members of Sarbecovirus clade 1. Phylogenetic trees were built using PhyML [35]. Spike protein sequences from human pathogenic CoVs are indicated in bold. Host species are shown for some of the nodes as dark silhouettes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)