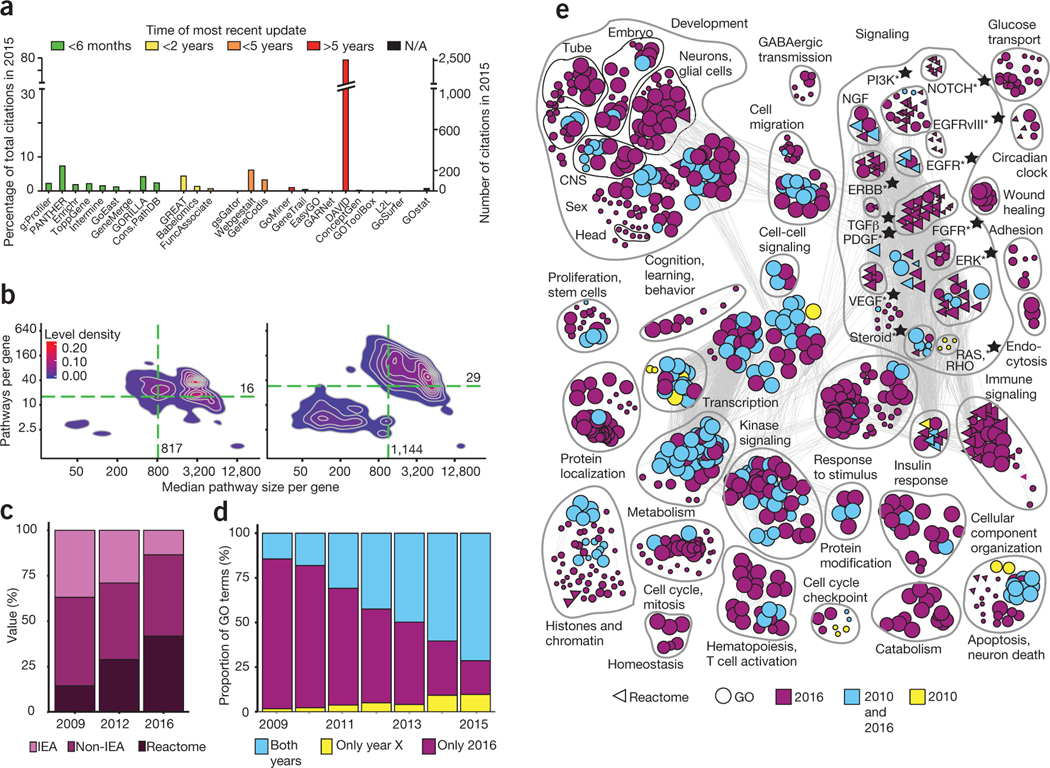
Figure 1 |.
Outdated pathway analysis resources strongly affect practical genomic analysis and literature. (a) The majority of public software tools for pathway enrichment analysis use outdated gene annotations, and the majority of surveyed papers published in 2015 used annotations that were more than five years old. (b) Density plots showing the evolution of pathway knowledge (GO + Reactome) between 2009 (left) and 2016 (right). The values for the median gene are indicated by green dashed lines. The bottom left group in the 2016 plot corresponds to Reactome pathways. (c) Gene annotation quality is improving rapidly as manually curated Reactome annotations are becoming more frequent and fewer genes in GO are IEA. (d) Pathway enrichment analysis of frequently mutated GBM genes showing the proportion of results missed in outdated GO annotations. Each bar compares annotations from a given year to 2016 annotations. (e) Enrichment map of frequently mutated GBM pathways and processes according to gene annotations from 2010 and 2016. Three-quarters of current findings are missed in out-of-date analyses (purple). Nodes represent processes and pathways, and edges connect nodes with many shared genes. Stars indicate clinically actionable pathways.